Abstract
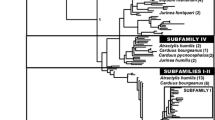
A family of satellite DNA is analyzed in seven ant species from the genus Aphaenogaster. This satellite DNA is organized as tandemly repeated sequences with a consensus sequence of 160 bp in length. The sampled sequences show a high similarity and belong to the same family of satellite DNA. However in Aphaenogaster spinosa, two types of repeat clearly differentiated have been found. Phylogenetic analyses using satellite DNA show that sequences do not cluster in a species-specific way, with one exception. Concretely, the second type of repeats of A. spinosa (APSP-II) which constitutes a new satellite DNA subfamily. The obtained results with satellite DNA are compared with those obtained using mitochondrial and nuclear DNA to determine the correlation between evolution of satellite DNA and phylogenetic relationships among the analyzed ants. The high interspecific similarity for the satellite DNA seems not to be in concordance with the concerted evolution pattern, commonly accepted to explain the evolution of satellite DNA. However, the accumulated data suggest that evolution of satellite DNA in ants follows the concerted evolution pattern but that this process is slow in relation with other organisms, probably due to the eusociality and haplodiploidy of these insects.



Similar content being viewed by others
References
Betrán, E., Rozas, J., Navarro, A., & Barbadilla, A. (1997). The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics, 146, 89–99.
Blaimer, B. B., Brady, S. G., Schultz, T. R., Lloyd, M. W., Fisher, B. L., & Ward, P. S. (2015). Phylogenomic methods outperform traditional multi-locus approaches in resolving deep evolutionary history: a case study of formicine ants. BMC Evolutionary Biology, 15, 271. doi:10.1186/s12862-015-0552-5.
Boer, P. (2013). Revision of the European ants of the Aphaenogaster testaceopilosa-group (Hymenoptera: Formicidae). Tijdschrift Voor Entomologie, 156, 57–93. doi:10.1163/22119434-00002022.
Bolton, B. (1982). Afrotropical species of the myrmicine ant genera Cardiocondyla, Leptothorax, Melissotarsus, Messor and Cataulacus (Formicidae). Bulletin of the British Museum Natural History (Entomology), 4, 307–370.
Bolton, B. (2016). New general catalogue of the ants of the world, including a synopsis of taxonomic publications on Formicidae. VERSION: 3 MAY 2016. http://www.antwiki.org/wiki/Species_Accounts, December 2016.
Brady, S. G., Schultz, T. R., Fisher, B. L., & Ward, P. S. (2006). Evaluating alternative hypotheses for the early evolution and diversification of ants. Proceedings of the National Academy of Sciences of the United States of America, 103, 18172–18177. doi:10.1073/pnas.0605858103.
Chiotis, M., Jermiin, L. S., & Crozier, R. H. (2000). A molecular framework for the phylogeny of the ant subfamily Dolichoderinae. Molecular Phylogenetics and Evolution, 17, 108–116. doi:10.1006/mpev.2000.0821.
Dover, G. (2002). Molecular drive. Trends in Genetics, 18, 587–589. doi:10.1016/S0168-9525(02)02789-0.
Dowton, M., & Austin, A. D. (1998). Phylogenetic relationships among the microgastroid wasps (Hymenoptera: Braconidae): combined analysis of 16S and 28S rDNA genes and morphological data. Molecular Phylogenetics and Evolution, 10, 354–366. doi:10.1006/mpev.1998.0533.
Dowton, M., & Austin, A. D. (2001). Simultaneous analysis of 16S, 28S, COI and morphology in the Hymenoptera: Apocrita, evolutionary transitions among parasitic wasp. Biological Journal of the Linnean Society, 74, 87–111. doi:10.1006/bijl.2001.0577.
Espadaler, X. (1979). Una nueva hormiga de la Península Ibérica (Hymenoptera, Formicidae). Miscelanea Zoologica, 5, 77–81.
Feldhaar, H., Fiala, B., Gadau, J., Mohamed, M., & Maschwitz, U. (2003). Molecular phylogeny of Crematogaster subgenus Decacrema ants (Hymenoptera: Formicidae) and the colonization of Macaranga (Euphorbiaceae) trees. Molecular Phylogenetics and Evolution, 27, 441–452. doi:10.1016/S1055-7903(02)00449-9.
Fry, K., & Salser, W. (1977). Nucleotide sequences of HS-a satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell, 12, 1069–1084. doi:10.1016/0092-8674(77)90170-2.
Gillespie, J. J., Johnston, J. S., Cannone, J. J., & Gutell, R. R. (2006). Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Molecular Biology, 15, 657–686. doi:10.1111/j.1365-2583.2006.00689.x.
Huang, Y. C., Lee, C. C., Kao, C. Y., Chang, N. C., Lin, C. C., Shoemaker, D., & Wang, J. (2016). Evolution of long centromeres in fire ants. BMC Evolutionary Biology, 16, 189. doi:10.1186/s12862-016-0760-7.
Janda, M., Folková, D., & Zrzavý, J. (2004). Phylogeny of Lasius ants based on mitochondrial DNA and morphology, and the evolution of social parasitism in the Lasiini (Hymenoptera: Formicidae). Molecular Phylogenetics and Evolution, 33, 595–614. doi:10.1016/j.ympev.2004.07.012.
Jukes, T. H., & Cantor, C. R. (1969). Evolution of protein molecules. In H. N. Munro (Ed.), Mammalian protein metabolism (pp. 21–132). New York: Academic Press.
Kuhn, G. C., Küttler, H., Moreira-Filho, O., & Heslop-Harrison, J. S. (2012). The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Molecular Biology and Evolution, 29, 7–11. doi:10.1093/molbev/msr173.
Librado, P., & Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. doi:10.1093/bioinformatics/btp187.
Lorite, P., Carrillo, J. A., Aguilar, J. A., & Palomeque, T. (2004a). Isolation and characterization of two families of satellite DNA with repetitive units of 135 bp and 2.5 kb in the ant Monomorium subopacum (Hymenoptera, Formicidae). Cytogenetic and Genome Research, 105, 83–92. doi:10.1159/000078013.
Lorite, P., Carrillo, J. A., Tinaut, A., & Palomeque, T. (2002a). Comparative study of satellite DNA in ants of the Messor genus (Hymenoptera, Formicidae). Gene, 297, 113–122. doi:10.1016/S0378-1119(02)00875-2.
Lorite, P., Carrillo, J. A., Tinaut, A., & Palomeque, T. (2004b). Evolutionary dynamics of satellite DNA in species of the genus Formica (Hymenoptera, Formicidae). Gene, 332, 159–168. doi:10.1016/j.gene.2004.02.049.
Lorite, P., García, M. F., & Palomeque, T. (1999). Satellite DNA in the ant Messor structor. Genome, 42, 881–886. doi:10.1139/g99-018.
Lorite, P., Renault, S., Rouleux-Bonnin, F., Bigot, S., Periquet, G., & Palomeque, T. (2002b). Genomic organization and transcription of satellite DNA in the ant Aphaenogaster subterranea (Hymenoptera, Formicidae). Genome, 45, 609–616. doi:10.1139/g02-022.
Luchetti, A., Cesari, M., Carrara, G., Cavicchi, S., Passamonti, M., Scali, V., & Mantovani, B. (2003). Unisexuality and molecular drive: Bag320 sequence diversity in Bacillus taxa (insecta phasmatodea). Journal of Molecular Evolution, 56, 587–596. doi:10.1007/s00239-002-2427-9.
Luchetti, A., Marini, M., & Mantovani, B. (2006). Non-concerted evolution of the RET76 satellite DNA family in Reticulitermes taxa (Insecta, Isoptera). Genetica, 128, 123–132. doi:10.1007/s10709-005-5540-z.
Mantovani, B., Tinti, F., Bachmann, L., & Scalli, V. (1997). The Bag320 satellite DNA family in Bacillus stick insects (Phasmatodea): different rates of molecular evolution of highly repetitive DNA in bisexual and parthenogenetic taxa. Molecular Biology and Evolution, 14, 1197–1205.
Mestrović, N., Plohl, M., Mravinac, B., & Ugarković, D. (1998). Evolution of satellite DNAs from the genus Palorus—experimental evidence for the “library” hypothesis. Molecular Biology and Evolution, 15, 1062–1068. doi:10.1093/oxfordjournals.molbev.a026005.
Moreau, C. S., Bell, C. D., Vila, R., Archibald, S. B., & Pierce, N. E. (2006). Phylogeny of the ants: diversification in the age of angiosperms. Science, 312, 101–104. doi:10.1126/science.1124891.
Muñoz-López, M., Palomeque, T., Carrillo, J. A., Pons, J., Tinaut, A., & Lorite, P. (2012). A new taxonomic status for Iberoformica (Hymenoptera, Formicidae) based on the use of molecular markers. Journal of Zoological Systematics and Evolutionary Research, 50, 30–37. doi:10.1111/j.1439-0469.2011.00649.x.
Navajas-Pérez, R., Quesada del Bosque, M. E., & Garrido-Ramos, M. A. (2009). Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Molecular Genetics and Genomics, 282, 395–406. doi:10.1007/s00438-009-0472-4.
Nei, M. (1987). Molecular evolutionary genetics. New York: Columbia University Press.
Nei, M., & Miller, J. C. (1990). A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics, 125, 873–879.
Palomeque, T., & Lorite, P. (2008). Satellite DNA in insects—a review. Heredity, 100, 564–573. doi:10.1038/hdy.2008.24.
Palomeque, T., Sanllorente, O., Maside, X., Vela, J., Mora, P., Torres, M. I., Periquet, G., & Lorite, P. (2015). Evolutionary history of the Azteca-like mariner transposons and their host ants. Naturwissenschaften, 102, 44. doi:10.1007/s00114-015-1294-3.
Pavlek, M., Gelfand, Y., Plohl, M., & Meštrović, N. (2015). Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Research, 22, 387–401. doi:10.1093/dnares/dsv021.
Pérez-Gutiérrez, M. A., Suárez-Santiago, V. N., López-Flores, I., Romero, A. T., & Garrido-Ramos, M. A. (2012). Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Molecular Biology, 78, 19–29. doi:10.1007/s11103-011-9848-z.
Plohl, M., Meštrović, N., & Mravinac, B. (2012). Satellite DNA evolution. Genome Dynamics, 7, 126–152. doi:10.1159/000337122.
Plohl, M., Petrović, V., Luchetti, A., Ricci, A., Satović, E., Passamonti, M., & Mantovani, B. (2010). Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity, 104, 543–551. doi:10.1038/hdy.2009.141.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 1, 1572–1574.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425.
Santschi, F. (1932). Etudes sur quelques Attomyrma palearctiques. Bulletin Société Entomologique Suisse, 15, 338–346.
Schulz, A. (1994). Aphaenogaster graeca nova species (Hym: Formicidae) aus dem Olyp-Gebirge (Griechenland) und eine Gliederung der Gattung Aphaenogaster. Beiträge Zur Entomologie, 44, 417–429.
Sharma, A., Wolfgruber, T. K., & Presting, G. G. (2013). Tandem repeats derived from centromeric retrotransposons. BMC Genomics, 14, 142. doi:10.1186/1471-2164-14-142.
Stephan, W. (1997). Tandemly repeated non-coding DNA sequences. In: Meyers, R.A. (Ed.). The encyclopedia of molecular biology and molecular medicine Volume 6, pp. 1–10. Verlagsgesellschaft, Weinheim.
Strachan, T., Webb, D., & Dover, G. A. (1985). Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO Journal, 4, 1701–1708.
Talbert, P. B., & Henikoff, S. (2010). Centromeres convert but don’t cross. PLoS Biology, 8, e1000326. doi:10.1371/journal.pbio.1000326.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA 6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. doi:10.1093/molbev/mst197.
Tinaut, A. (1985). Descripción del macho de Aphaenogaster cardenai Espadaler, 1981 (Hymenoptera, Formicidae). Miscellania Zoologica, 9, 245–249.
Ward, P. S., Brady, S. G., Fisher, B. L., & Schultz, T. R. (2015). The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Systematic Entomology, 40, 61–81. doi:10.1111/syen.12090.
Wheeler, W. M. (1915). The ants of the Baltic Amber. Schriften der Physikalisch-Ökonomischen Gesellschaft Zu Königsberg, 55, 1–142.
Acknowledgements
This work was supported by the Spanish Junta de Andalucía (through the programs “Ayudas a Grupos de Investigación,” Group BIO220, and by the Spanish Ministerio de Ciencia e Innovación (through project CGL2011-23841, co-funded by the European Regional Development Fund). We thank Dr. Alain Lenoir from University of Tours (France) for providing us of the A. spinosa sample used in the present paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lorite, P., Muñoz-López, M., Carrillo, J. et al. Concerted evolution, a slow process for ant satellite DNA: study of the satellite DNA in the Aphaenogaster genus (Hymenoptera, Formicidae). Org Divers Evol 17, 595–606 (2017). https://doi.org/10.1007/s13127-017-0333-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-017-0333-7