Abstract
Spot blotch disease of wheat caused by Bipolaris sorokiniana Boerma (Sacc.) is an emerging problem in South Asian countries. Whole genome of a highly virulent isolate of B. sorokiniana BS112 (BHU, Uttar Pradesh; Accession no. GCA_004329375.1) was sequenced using a hybrid assembly approach. Secreted proteins, virulence gene(s), pathogenicity-related gene(s) were identified and the role of ToxA gene present in this genome, in the development of disease was recognized. ToxA gene (535 bp) was analyzed and identified in the genome of B. sorokiniana (BS112) which revealed 100% homology with the ToxA gene of Pyrenophora tritici repentis (Accession no. MH017419). Furthermore, ToxA gene was amplified, sequenced and validated in 39 isolates of B. sorokiniana which confirmed the presence of ToxA gene in all the isolates taken for this study. All ToxA sequences were submitted in NCBI database (MN601358-MN601396). As ToxA gene interacts with Tsn1 gene of host, 13 wheat genotypes were evaluated out of which 5 genotypes (38.4%) were found to be Tsn1 positive with more severe necrotic lesions compared to Tsn1-negative wheat genotypes. In vitro expression analysis of ToxA gene in the pathogen B. sorokiniana using qPCR revealed maximum upregulation (14.67 fold) at 1st day after inoculation (DAI) in the medium. Furthermore, in planta expression analysis of ToxA gene in Tsn1-positive and Tsn1-negative genotypes, revealed maximum expression (7.89-fold) in Tsn1-positive genotype, Agra local at 5th DAI compared to Tsn1-negative genotype Chiriya 7 showing minimum expression (0.048-fold) at 5th DAI. In planta ToxA–Tsn1 interaction studies suggested that spot blotch disease is more severe in Tsn1-positive genotypes, which will be helpful in better understanding and management of spot blotch disease of wheat.








Similar content being viewed by others
Data availability
Data are available in a publically accessible repository. The data presented in this study are openly available at https://www.ncbi.nlm.nih.gov.
References
Adlakha KL, Wilcoxson HD, Raychaudhury SP (1984) Resistance of wheat to leaf spot caused by B. sorokiniana. Plant Dis 68:320–321. https://doi.org/10.1094/PD-68-320
Aggarwal R, Das S, Jahani M, Singh D (2008) Histopathology of spot blotch disease of wheat caused by B. sorokiniana (Teleomorph: Cochliobolus sativus). Acta Phytopathol Entomol Hung 43(1):23–30. https://doi.org/10.1556/APhyt.43.2008.1.3
Aggarwal R, Singh VB, Gurjar MS, Gupta S, Srinivas P (2009) Intraspecific variations in Indian isolates of B. sorokiniana infecting wheat based on morphological, pathogenic and molecular characters. Indian Phytopath 62(4):449–460
Aggarwal R, Singh VB, Shukla R, Gurjar MS, Gupta S, Sharma TR (2010) URP-based DNA fingerprinting of B. sorokiniana isolates causing spot blotch of wheat. J Phytopathol 158:210–216. https://doi.org/10.1111/j.1439-0434.2009.01603.x
Aggarwal R, Sharma S, Singh K, Gurjar MS, Saharan MS, Gupta S, Bashyal BM, Gaikwad K (2019) First draft genome sequence of wheat spot blotch pathogen B. sorokiniana BS_112 from India, obtained using hybrid assembly. Microbiol Resour Announc 8(38):e00308-19. https://doi.org/10.1128/mra.00308-19
Alam KB, Banu SP, Shaheed MA (1998) The occurrence and significance of spot blotch disease in Bangladesh. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Proc int workshop on helminthosporium disease of wheat: spot blotch and tan spot. CIMMYT, El Batan, pp 63–66
Altschul SF, Gish W et al (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Antoni EA, Rybak K, Tucker MP, Hane JK, Solomon PS, Drenth A, Shankar M, Oliver RP (2010) Ubiquity of ToxA and absence of ToxB in Australian populations of P. tritici-repentis. Austral Plant Pathol 39:63–68. https://doi.org/10.1071/AP09056
Aylward J, Steenkamp ET, Dreyer LL, Roets F, Wingfield BD, Wingfield MJ (2017) A plant pathology perspective of fungal genome sequencing. IMA Fungus 8(1):1–15. https://doi.org/10.5598/imafungus.2017.08.01.01
Bryant JM, Baumgarten S, Lorthiois A, Scheidig-Benatar C, Claës A, Scherf A (2018) De Novo genome assembly of a Plasmodium falciparum NF54 clone using single- molecule real-time sequencing. Genome Announc 6(5):e01479-17. https://doi.org/10.1128/genomeA.01479-17
Chaurasia S, Chand R, Joshi AK (2000) Relative dominance of Alternaria triticina Pras. et Prab and B. sorokiniana (Sacc.) Shoemaker, in different growth stages of wheat (T. aestivum L.). J Plant Dis Prot 107:176–181. https://www.jstor.org/stable/43226857. Accessed 3 Nov 2021
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. https://doi.org/10.1155/2008/619832
Faris JD, Zhang Z, Lu H et al (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA 107(30):13544–13549. https://doi.org/10.1073/pnas.1004090107
Figueroa M, Hammond-Kosack KE, Solomon PS (2018) A review of wheat diseases-a field perspective. Mol Plant Pathol 19(6):1523–1536. https://doi.org/10.1111/mpp.12618
Finn RD, Bateman A, Clements J, Coggill P et al (2014) Pfam: the protein families database. Nucleic Acids Res 42:222–230. https://doi.org/10.1093/nar/gkt1223
Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Rasmussen JB, Solomon PS, McDonald BA, Oliver RP (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38:953–956. https://doi.org/10.1038/ng1839
Friesen TL, Holmes DJ, Bowden RL, Faris JD (2018) ToxA is present in the U.S. B. sorokiniana population and is a significant virulence factor on wheat harboring Tsn1. Plant Dis 102(12):2446–2452. https://doi.org/10.1094/pdis-03-18-0521-re
Gayad SK (1961) Production of symptoms of barley leaf spot disease by cultural filtrates of Helminthosporium sativum. Nature 191:725–726. https://doi.org/10.1038/191725b0
Gupta P, Vasistha N, Aggarwal R, Joshi A (2017) Biology of B. sorokiniana (syn. Cochliobolus sativus) in genomics era. J Plant Biochem Biotechnol 27(10):123–138. https://doi.org/10.1007/s13562-017-0426-6
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Collier CJA, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(2):85–87. https://doi.org/10.1093/nar/gkm259
Jahani M, Aggarwal R, Gupta S, Sharma S, Dureja P (2014) Purification and characterization of a novel toxin from B. sorokiniana, causing spot blotch of wheat and analysis of variability in the pathogen. Cereal Res Commun 42(2):252–261. https://doi.org/10.1556/CRC.2013.0053
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res D1(45):353–361. https://doi.org/10.1093/nar/gkw1092
Krueger F (2015) Trim galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinform 516:517
Kuan CS, Yew SM, Toh YF, Chan CL et al (2015) Dissecting the fungal biology of Bipolaris papendorfii: from phylogenetic to comparative genomic analysis. DNA Res 22(3):219–232. https://doi.org/10.1093/dnares/dsv007
Kumar J, Schafer P, Huckelhoven R, Langen G, Baltruschat H, Stein E, Nagarajan S, Kogel KH (2002) B. sorokiniana, a cereal pathogen of global concern: cytological and molecular approaches towards better control. Mol Plant Pathol 3:185–195. https://doi.org/10.1046/j.1364-3703.2002.00120.x
Kumar S, Roder MS, Tripathi SB, Kumar S, Chand R, Joshi AK, Kumar U (2015) Mendelization and fine mapping of a bread wheat spot blotch disease resistance QTL. Mol Breed 35:218. https://doi.org/10.1007/s11032-015-0411-5
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lam MQ, Oates NC, Thevarajoo S, Tokiman L, Goh KM, Mason SJM, Bruce NC, Chong CS (2020) Genomic analysis of a lignocellulose degrading strain from the underexplored genus Meridianimaribacter. Genomics 112(1):952–960. https://doi.org/10.1016/j.ygeno.2019.06.011
Livak KJ, Thomas S (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCt method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Manamgoda DS, Rossman AY, Castlebury LA, Crous PW, Madrid H, Chukeatirote E, Hyde KD (2014) The genus bipolaris. Stud Mycol 79:221–288. https://doi.org/10.1016/j.simyco.2014.10.002
Maraite H, Zinno TDI, Longree H, Daumerie V, Dueveiller E (1998) Fungi association with foliar blight of wheat areas. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Helminthosophism diseases of wheat: spot blotch and tan spot. CIMMYT, pp 293–300
McDonald MC, Oliver RP, Friesen TL, Brunner PC, McDonald BA (2013) Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol 199:241–251. https://doi.org/10.1111/nph.12257
McDonald MC, Ahren D, Simpfendorfer S, Milgate A, Solomon PS (2018) The discovery of the virulence gene ToxA in the wheat and barley pathogen B. sorokiniana. Mol Plant Pathol 19(2):432–439. https://doi.org/10.1111/mpp.12535
McDonald MC, Taranto AP, Hill E, Schwessinger B, Liu Z, Simpfendorfer S, Milgate A, Solomon PS (2019) Transposon-mediated horizontal transfer of the host-specific virulence protein ToxA between three fungal wheat pathogens. mBio 10(5):e01515-19. https://doi.org/10.1128/mBio.01515-19
Moolhuijzen PM, See PT, Oliver RP, Moffat CS (2018) Genomic distribution of a novel P. tritici-repentis ToxA insertion element. PLoS One 13(10):0206586. https://doi.org/10.1371/journal.pone.0206586
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4325. https://doi.org/10.1093/nar/8.19.4321
Nagarajan S, Kumar J (1998) Foliar blights of wheat in India: germplasm improvement and future challenges for sustainable high yielding wheat production. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Proc Int Workshop Helminthosporium Diseases of Wheat: Spot Blotch and Tan Spot, 9–14 February 1997. CIMMYT, El Batan, pp 52–58
Navathe S, Yadav PS, Chand R, Mishra VK, Vasistha NK, Meher PK, Joshi AK, Gupta PK (2020) ToxA–Tsn1 interaction for spot blotch susceptibility in Indian wheat: an example of inverse gene-for-gene relationship. Plant Dis 104(1):71–78. https://doi.org/10.1094/PDIS-05-19-1066-RE
Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA et al (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog 8(12):e1003037. https://doi.org/10.1371/journal.ppat.1003037
Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM (2016) Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. https://doi.org/10.1186/s13059-016-0997-x
Peterson TN, Brunak S, Von HG, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Ramadas S, Kumar KTM, Pratap SG (2020) Wheat Production in India: trends and prospects. In: Shah F, Khan Z, Iqbal A, Turan M, Olgun M (eds) Recent advances in grain crops research. Intechopen
Sahoo N (2017) Sequence Base-calling through Albacore software: A part of the Oxford Nanopore Technology. Dissertation, Orissa University of Agriculture and Technology
Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. https://doi.org/10.1093/nar/gki458
Stukenbrock EH, McDonald BA (2007) Geographical variation and positive diversifying selection in the host-specific toxin SnToxA. Mol Plant Pathol 8:321–332. https://doi.org/10.1111/j.1364-3703.2007.00396.x
Svobodova L, Hanzalová A, Kucera L (2010) Expansion and variability of the Ptr Tox A gene on populations of P. tritici-repentis and P. teres. J Plant Pathol 92:729–735
Van Ginkel M, Rajaram S (1993) Breeding for durable resistance to diseases in wheat: an additional perspective. In: Jacobes T, Parlevliet JE (eds) Durability of disease resistance. Kluwer Academic Publishers, Dordrecht, pp 259–272
Wiese MV (1998) Compendium of wheat diseases. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Proc int workshop helminthosporium disease of wheat: spot blotch and tan spot. 9–14 Feb. 1997. CIMMYT, El Batan, pp 114–118
Wu L, He X, Lozano N et al (2021) ToxA, a significant virulence factor involved in wheat spot blotch disease, exists in the Mexican population of B. sorokiniana. Trop Plant Pathol 46(2):201–206. https://doi.org/10.1007/s40858-020-00391-4
Zillinsky FJ (1983) In: common diseases of small grain cereals, a guide to identification. CIMMYT, p 141
Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA (2013) The MaSuRCA genome assembler. Bioinformatics 29(21):2669–2677. https://doi.org/10.1093/bioinformatics/btt476
Acknowledgements
Authors are highly thankful to the ICAR-Consortium Research Platform (CRP) on Genomics (ICAR-G/CRP-Genomics/2015-2720/IARI-12-151) for funding this work. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We are very thankful to the Director and Joint Director (Research) of the ICAR-Indian Agricultural Research Institute, New Delhi, the Head, Division of Plant Pathology, ICAR-IARI, New Delhi, and the Director of ICAR-NIPB, New Delhi for providing facilities.
Author information
Authors and Affiliations
Contributions
RA, SA and SS conceived and designed the experiment and wrote the manuscript; RA, BMB, MSG and MSS contributed in corrections and suggestions; ARR and SS helped in performing bioinformatics analysis; RA, SA and PJ analyzed the data and conducted the wet lab experiments. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2022_3213_MOESM3_ESM.xls
Supplementary file3 (XLS 323 KB) Annotation of fungal hybrid assembly in terms of GO proteins – Biological Process, Cellular Component, Molecular Function of B. sorokiniana

13205_2022_3213_MOESM4_ESM.jpg
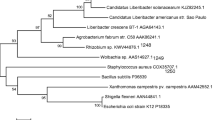
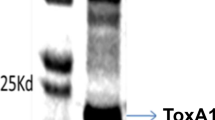
Supplementary file4 (JPG 1290 KB) a PCR amplification profile of 39 isolates of B. sorokiniana harbouring ToxA (535 bp amplicon) gene. M (100 bp bioprep marker), 1-BS1, 2-BS3, 3-BS4, 4-BS6, 5-BS11, 6-BS14, 7-BS27, 8-BS29, 9-BS30, 10-BS32, 11-BS50, 12-BS54, 13-BS65, 14-BS66, 15-BS68, 16-BS69, 17-BS72, 18-BS75, 19-BS88, 20-BS91, 21-BS94, 22-BS112, 23-BS124, 24-BS125, 25-BS126, 26-BS127, 27-BS128, 28-BS129, 29-BS130, 30-BS131, 31-BS132, 32-BS133, 33-BS134, 34-BS135, 35-BS136, 36-BS137, 37-BS138, 38-BS139, 39-BS140. b Phylogenetic tree constructed based on ToxA gene sequences of different isolates of B. sorokiniana, P. tritici-repentis, P. avenaria and P. nodorum. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA X


13205_2022_3213_MOESM6_ESM.jpg
Supplementary file6 (JPG 9362 KB) Alignment of nucleotide sequences of B. sorokiniana ToxA gene from India (BSToxA) and Australia (AusBSToxA1 and AusBSToxA2) aligned to P. tritici-repentis (ptr_ToxA) and P. nodorum (Sn4_ToxA)

13205_2022_3213_MOESM7_ESM.jpg
Supplementary file7 (JPG 5644 KB) Pathogenicity assay symptoms on different wheat genotypes inoculated with BS112. 1-WH 542, 2-WL 711, 3-Agra Local, 4-Suzoe, 5- HD 29, 6- PBW 343, 7-HD 3086, 8-Chiriya 7, 9-HD 30, 10-Milan, 11-HD 2329, 12- Ning 8139, 13-HD 2967
Rights and permissions
About this article
Cite this article
Aggarwal, R., Agarwal, S., Sharma, S. et al. Whole-genome sequence analysis of Bipolaris sorokiniana infecting wheat in India and characterization of ToxA gene in different isolates as pathogenicity determinants. 3 Biotech 12, 151 (2022). https://doi.org/10.1007/s13205-022-03213-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03213-3