Abstract
Background
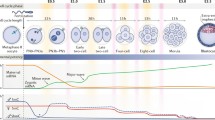
Dynamic chromatin reorganization occurs during two waves of cell lineage specification process, blastocyst formation and gastrulation, to generate distinct cell types. Epigenetic defects have been associated with severe developmental defects and diseases. How epigenetic remodeling coordinates the two lineage specification waves is becoming uncovered, benefiting from the development and application of new technologies including low-input or single-cell epigenome analysis approached in the past few years.
Objective
In this review, we aim to highlight the most recent findings on epigenetic remodeling in cell lineage specification during blastocyst formation and gastrulation.
Methods
First, we introduce how DNA methylation dynamically changes in blastocyst formation and gastrulation and its function in transcriptional regulation lineage-specific genes. Then, we discuss widespread remodeling of histone modification at promoters and enhancers in orchestrating the trajectory of cell lineage specification. Finally, we review dynamics of chromatin accessibility and 3D structure regulating developmental gene expression and associating with specific transcription factor binding events at stage specific manner. We also highlight the key questions that remain to be answered to fully understand chromatin regulation and reorganization in lineage specification.
Conclusion
Here, we summarize the recent advances and discoveries on epigenetic reorganization and its roles in blastocyst formation and gastrulation, and how it cooperates with the lineage specification, painting from global sequencing data from mouse in vivo tissues.

Similar content being viewed by others
References
Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng DY et al (2015) Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521:366–370
Andersson R, Sandelin A (2020) Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet 21:71–87
Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, Imaz-Rosshandler I, Lohoff T, Xiang YL, Hanna CW et al (2019) Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576:487–491
Arnold SJ, Robertson EJ (2009) Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10:91–103
Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M et al (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–538
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395
Beagrie RA, Scialdone A, Schueler M, Kraemer DCA, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR et al (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543:519–524
Bernstein BE, Mikkelsen TS, Xie XH, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K et al (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326
Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8:1499–1504
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21
Bowman GD, Poirier MG (2015) Post-translational modifications of histones that influence nucleosome dynamics. Chem Rev 115:2274–2295
Burton A, Torres-Padilla ME (2014) Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15:722–734
Calo E, Wysocka J (2013) Modification of enhancer chromatin: what, how, and why? Mol Cell 49:825–837
Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43:559–599
Chazaud C, Yamanaka Y (2016) Lineage specification in the mouse preimplantation embryo. Development 143:1063–1074
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA et al (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107:21931–21936
Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P (2010) Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS ONE 5:e9150
Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF (2014) Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141:526–537
Di Croce L, Helin K (2013) Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20:1147–1155
Dileep V, Rivera-Mulia JC, Sima J, Gilbert DM (2015) Large-scale chromatin structure-function relationships during the cell cycle and development: insights from replication timing. Cold Spring Harb Symp Quant Biol 80:53–63
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380
Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W et al (2015) Chromatin architecture reorganization during stem cell differentiation. Nature 518:331–336
Ea V, Baudement MO, Lesne A, Forne T (2015) Contribution of topological domains and loop formation to 3D chromatin organization. Genes 6:734–750
Fergusonsmith AC, Sasaki H, Cattanach BM, Surani MA (1993) Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362:751–755
Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K (2017) Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544:110–114
Fortin JP, Hansen KD (2015) Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol 16:180
Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D et al (2018) Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173:248–259
Greenberg MVC, Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20:590–607
Heinz S, Romanoski CE, Benner C, Glass CK (2015) The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16:144–154
Iguchiariga SMM, Schaffner W (1989) CpG methylation of the cAMP-responsive enhancer promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 3:612–619
Inoue A, Zhang Y (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334:194–194
Iqbal K, Jin SG, Pfeifer GP, Szabo PE (2011) Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA 108:3642–3647
Jambhekar A, Dhall A, Shi Y (2019) Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 20:625–641
Jeong H, Mendizabal I, Berto S, Chatterjee P, Layman T, Usui N, Toriumi K, Douglas C, Singh D, Huh I et al (2021) Evolution of DNA methylation in the human brain. Nat Commun 12:2021
Ji Z, He LZ, Rotem A, Janzer A, Cheng CS, Regev A, Struhl K (2018) Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation. Nat Commun 9:2068
Klemm SL, Shipony Z, Greenleaf WJ (2019) Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20:207–220
Kohli RM, Zhang Y (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502:472–479
Kojima Y, Tam OH, Tam PP (2014) Timing of developmental events in the early mouse embryo. Semin Cell Dev Biol 34:65–75
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Lawrence M, Daujat S, Schneider R (2016) Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32:42–56
Li E, Zhang Y (2014) DNA methylation in mammals. Cold Spring Harb Perspect Biol 6:a019133
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293
Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD et al (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905
Liu XY, Wang CF, Liu WQ, Li JY, Li C, Kou XC, Chen JY, Zhao YH, Gao HB, Wang H et al (2016) Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537:558–562
Lu ZF, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ (2017) Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45:e41
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349
Moore LD, Le T, Fan GP (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38
Niu YY, Sun NQ, Li C, Lei Y, Huang ZH, Wu J, Si CY, Dai X, Liu CY, Wei JK et al (2019) Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science 366:eaaw5754
Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA (2018) Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci USA 115:E6697–E6706
Ong CT, Corces VG (2012) Enhancers: emerging roles in cell fate specification. EMBO Rep 13:423–430
Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong CT, Hookway TA, Guo CY, Sun YH et al (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153:1281–1295
Pijuan-Sala B, Wilson NK, Xia J, Hou X, Hannah RL, Kinston S, Calero-Nieto FJ, Poirion O, Preissl S, Liu F et al (2020) Single-cell chromatin accessibility maps reveal regulatory programs driving early mouse organogenesis. Nat Cell Biol 22:487–497
Posfai E, Tam OH, Rossant J (2014) Mechanisms of pluripotency in vivo and in vitro. Curr Top Dev Biol 107:1–37
Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al (2014) A 3d map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680
Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293:1089–1093
Ren B, Yue F (2015) Transcriptional enhancers: bridging the genome and phenome. Cold Spring Harb Symp Quant Biol 80:17–26
Rossant J, Tam PP (2004) Emerging asymmetry and embryonic patterning in early mouse development. Dev Cell 7:155–164
Rowley MJ, Corces VG (2018) Organizational principles of 3D genome architecture. Nat Rev Genet 19:789–800
Rubin AJ, Barajas BC, Furlan-Magaril M, Lopez-Pajares V, Mumbach MR, Howard I, Kim DS, Boxer LD, Cairns J, Spivakov M et al (2017) Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet 49:1522–1528
Ryba T, Hiratani I, Lu JJ, Itoh M, Kulik M, Zhang JF, Schulz TC, Robins AJ, Dalton S, Gilbert DM (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20:761–770
Schoenfelder S, Mifsud B, Senner CE, Todd CD, Chrysanthou S, Darbo E, Hemberger M, Branco MR (2018) Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages. Nat Commun 9:4189
Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB (2014) Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell 55:319–331
Smith ZD, Shi JT, Gu HC, Donaghey J, Clement K, Cacchiarelli D, Gnirke A, Michor F, Meissner A (2017) Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549:543–547
Stroud H, Su SC, Hrvatin S, Greben AW, Renthal W, Boxer LD, Nagy MA, Hochbaum DR, Kinde B, Gabel HW et al (2017) Early-life gene expression in neurons modulates lasting epigenetic states. Cell 171:1151–1164
Toenhake CG, Fraschka SAK, Vijayabaskar MS, Westhead DR, Heeringen SJ, Bartfai R (2018) Chromatin accessibility-based characterization of the gene regulatory network underlying Plasmodium falciparum blood-stage development. Cell Host Microbe 23:557-569.e9
Tsompana M, Buck MJ (2014) Chromatin accessibility: a window into the genome. Epigenet Chromatin 7:33
Wang A, Yue F, Li Y, Xie RY, Harper T, Patel NA, Muth K, Palmer J, Qiu YJ, Wang JZ et al (2015) Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16:386–399
Wang Y, Yuan P, Yan Z, Yang M, Huo Y, Nie Y, Zhu X, Qiao J, Yan L (2021) Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat Commun 12:1247
White MD, Plachta N (2020) Specification of the first mammalian cell lineages in vivo and in vitro. Cold Spring Harb Perspect Biol 12:a035634
Wu H, Zhang Y (2014) Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156:45–68
Wu JY, Huang B, Chen H, Yin QZ, Liu Y, Xiang YL, Zhang BJ, Liu BF, Wang QJ, Xia WK et al (2016) The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534:652–657
Xia WK, Xu JW, Yu G, Yao GD, Xu K, Ma XS, Zhang N, Liu BF, Li T, Lin ZL et al (2019) Resetting histone modifications during human parental-to-zygotic transition. Science 365:353–360
Xiang YL, Zhang Y, Xu QH, Zhou C, Liu BF, Du ZH, Zhang K, Zhang BJ, Wang XX, Gayen S et al (2020) Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat Genet 52:95–105
Xu Q, Xie W (2018) Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol 28:237–253
Yang XF, Hu BQ, Hou Y, Qiao YB, Wang R, Chen YY, Qian Y, Feng S, Chen J, Liu C et al (2018) Silencing of developmental genes by H3K27me3 and DNA methylation reflects the discrepant plasticity of embryonic and extraembryonic lineages. Cell Res 28:593–596
Yang XF, Hu BQ, Liao JY, Qiao YB, Chen YY, Qian Y, Feng S, Yu F, Dong J, Hou Y et al (2019) Distinct enhancer signatures in the mouse gastrula delineate progressive cell fate continuum during embryo development. Cell Res 29:911–926
Yang M, Tao X, Titus S, Zhao TH, Scott RT, Seli E (2020) Analysis of accessible chromatin landscape in the inner cell mass and trophectoderm of human blastocysts. Mol Hum Reprod 26:702–711
Zhang TY, Cooper S, Brockdorff N (2015) The interplay of histone modifications—writers that read. EMBO Rep 16:1467–1481
Zhang Y, Xiang YL, Yin QZ, Du ZH, Peng X, Wang QJ, Fidalgo M, Xia WK, Li YY, Zhao ZA et al (2018) Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat Genet 50:96–105
Zheng H, Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20:535–550
Zheng H, Huang B, Zhang BJ, Xiang YL, Du ZH, Xu QH, Li YY, Wang QJ, Ma J, Peng X et al (2016) Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 63:1066–1079
Zhou F, Wang R, Yuan P, Ren YX, Mao YU, Li R, Lian Y, Li JS, Wen L, Yan LY et al (2019) Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 572:660–664
Acknowledgements
We are grateful to the members in Lin & Luo Lab for critical discussion. This work was financially supported by grants from the National Key R&D Program of China (2018YFA0800100 and 2018YFA0800101 to C.L.; 2018YFA0800104 to H.F.), the National Natural Science Foundation of China (32030017 and 31970626 to C.L.) and the Natural Science Foundation of Jiangsu Province (BK20201273 to H.F.)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fang, H., Luo, Z. & Lin, C. Epigenetic reorganization during early embryonic lineage specification. Genes Genom 44, 379–387 (2022). https://doi.org/10.1007/s13258-021-01213-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01213-w