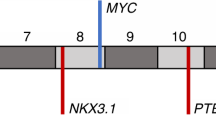
Abstract
The molecular cause of prostate cancer (PCa) is still unclear; however, its progression involves androgen, PI3K/Akt, and PTEN signaling, as cycle and apoptotic pathways. Alterations in oncogenes and tumor suppressor genes as PIK3CA, BRAF, KRAS and TP53 are not very common. Recently, somatic mutations have been discovered in relation to cancer progression mainly in genes such as PIK3CA; however, little data has been described in PCa. Nowadays genetic tools allow us to investigate multiple details about the biological heterogeneity of PCa, to better understand the mechanisms of disease progression and treatment resistance. Therefore, if the most relevant somatic mutations were included during screening, we could identify the best treatment for the right patient, bringing us closer to personalized medicine. The main objective of this article is to provide a review of the principal somatic mutations that appear to have a relevant role in hormonal cancers, like prostate cancer.

Similar content being viewed by others
References
Kral M, Rosinska V, Student V, Grepl M, Hrabec M, Bouchal J. Genetic determinants of prostate cancer: a review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2011;155(1):3–9.
Ostrander EA, Markianos K, Stanford JL. Finding prostate cancer susceptibility genes. Annu Rev Genomics Hum Genet. 2004;5:151–75.
Paolillo C, Londin E, Fortina P. Next generation sequencing in cancer: opportunities and challenges for precision cancer medicine. Scand J Clin Lab Invest Suppl. 2016;245:S84–91.
Aung KL, Board RE, Ellison G, Donald E, Ward T, Clack G, et al. Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J. 2010;4(1–4):11–21.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Fröhling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12(6):501–13.
Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–6.
Jackson BL, Grabowska A, Ratan HL. MicroRNA in prostate cancer: functional importance and potential as circulating biomarkers. BMC Cancer. 2014;14:930.
Lai CH, Huang SF, Liao CT, Chen IH, Wang HM, Hsieh LL. Clinical significance in oral cavity squamous cell carcinoma of pathogenic somatic mitochondrial mutations. PLoS One. 2013;8(6):e65578.
Lindberg J, Mills IG, Klevebring D, Liu W, Neiman M, Xu J, et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol. 2013;63(4):702–8.
Kloss-Brandstätter A, Schäfer G, Erhart G, Hüttenhofer A, Coassin S, Seifarth C, et al. Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am J Hum Genet. 2010;87(6):802–12.
van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF, et al. How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev Mutat Res. 2015;764:16–30.
INC MLS. The Prostate Core Mitomic Test™. 2016 [cited 2016 15 March]; Available from: http://mdnalifesciences.com/prostate-core-mitomic-test/.
Shull AY, Clendenning ML, Ghoshal-Gupta S, Farrell CL, Vangapandu HV, Dudas L, et al. Somatic mutations, allele loss, and DNA methylation of the Cub and Sushi Multiple Domains 1 (CSMD1) gene reveals association with early age of diagnosis in colorectal cancer patients. PLoS One. 2013;8(3):e58731.
Chaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhathat S. Somatic mutations of K-ras and BRAF in Thai colorectal cancer and their prognostic value. Asian Pac J Cancer Prev. 2013;14(1):329–32.
Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13.
Barbieri CE, Tomlins SA. The prostate cancer genome: perspectives and potential. Urol Oncol. 2014;32(1):53.e15–22.
Rubin MA, Girelli G, Demichelis F. Genomic correlates to the newly proposed grading prognostic groups for prostate cancer. Eur Urol. 2016;69(4):557–60.
Miyagi Y, Sasaki T, Fujinami K, Sano J, Senga Y, Miura T, et al. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010;23(11):1492–8.
Tapia-Laliena MA, Korzeniewski N, Hohenfellner M, Duensing S. High-risk prostate cancer: a disease of genomic instability. Urol Oncol. 2014;32(8):1101–7.
Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21(1):47–55.
Mehra R, Kumar-Sinha C, Shankar S, Lonigro RJ, Jing X, Philips NE, et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin Cancer Res. 2011;17(12):3924–32.
Mitchell T, Neal DE. The genomic evolution of human prostate cancer. Br J Cancer. 2015;113(2):193–8.
Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–7.
Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47(4):367–72.
Network CGAR. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–25.
Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–65.
Richardson RB. p53 mutations associated with aging-related rise in cancer incidence rates. Cell Cycle. 2013;12(15):2468–78.
Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008.
Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–11.
Morris EV, Cerundolo L, Lu M, Verrill C, Fritzsche F, White MJ, et al. Nuclear iASPP may facilitate prostate cancer progression. Cell Death Dis. 2014;5:e1492.
Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2(3):a001016.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28.
Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ, et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32(35):4130–8.
Huang L, Carney J, Cardona DM, Counter CM. Decreased tumorigenesis in mice with a Kras point mutation at C118. Nat Commun. 2014;5:5410.
Nodin B, Zendehrokh N, Sundström M, Jirström K. Clinicopathological correlates and prognostic significance of KRAS mutation status in a pooled prospective cohort of epithelial ovarian cancer. Diagn Pathol. 2013;8:106.
Yuan F, Shi M, Ji J, Shi H, Zhou C, Yu Y, et al. KRAS and DAXX/ATRX gene mutations are correlated with the clinicopathological features, advanced diseases, and poor prognosis in Chinese patients with pancreatic neuroendocrine tumors. Int J Biol Sci. 2014;10(9):957–65.
Wang XS, Shankar S, Dhanasekaran SM, Ateeq B, Sasaki AT, Jing X, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 2011;1(1):35–43.
Fu M, Zhang W, Shan L, Song J, Shang D, Ying J, et al. Mutation status of somatic EGFR and KRAS genes in Chinese patients with prostate cancer (PCa). Virchows Arch. 2014;464(5):575–81.
Reis ST, Timoszczuk LS, Pontes-Junior J, Viana N, Silva IA, Dip N, et al. The role of micro RNAs let7c, 100 and 218 expression and their target RAS, C-MYC, BUB1, RB, SMARCA5, LAMB3 and Ki-67 in prostate cancer. Clinics (Sao Paulo). 2013;68(5):652–7.
Cai H, Memarzadeh S, Stoyanova T, Beharry Z, Kraft AS, Witte ON. Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer. Cancer Res. 2012;72(18):4672–81.
Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci. 2013;110(37):E3506–15.
Blair BG, Wu X, Zahari MS, Mohseni M, Cidado J, Wong HY, et al. A phosphoproteomic screen demonstrates differential dependence on HER3 for MAP kinase pathway activation by distinct PIK3CA mutations. Proteomics. 2015;15(2–3):318–26.
Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94(4):455–9.
Gabelli SB, Echeverria I, Alexander M, Duong-Ly KC, Chaves-Moreira D, Brower ET, et al. Activation of PI3Kα by physiological effectors and by oncogenic mutations: structural and dynamic effects. Biophys Rev. 2014;6(1):89–95.
Hou W, Liu J, Chen P, Wang H, Ye BC, Qiang F. Mutation analysis of key genes in RAS/RAF and PI3K/PTEN pathways in Chinese patients with hepatocellular carcinoma. Oncol Lett. 2014;8(3):1249–54.
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–91.
Qiu W, Schönleben F, Li X, Ho DJ, Close LG, Manolidis S, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441–6.
Mullerad M, Hricak H, Kuroiwa K, Pucar D, Chen HN, Kattan MW, et al. Comparison of endorectal magnetic resonance imaging, guided prostate biopsy and digital rectal examination in the preoperative anatomical localization of prostate cancer. J Urol. 2005;174(6):2158–63.
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554.
Schönleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12(12):3851–5.
Tang KD, Ling MT. Targeting drug-resistant prostate cancer with dual PI3K/mTOR inhibition. Curr Med Chem. 2014;21(26):3048–56.
Wong HL, Peters U, Hayes RB, Huang WY, Schatzkin A, Bresalier RS, et al. Polymorphisms in the adenomatous polyposis coli (APC) gene and advanced colorectal adenoma risk. Eur J Cancer. 2010;46(13):2457–66.
Pećina-Slaus N, Majić Z, Musani V, Zeljko M, Cupić H. Report on mutation in exon 15 of the APC gene in a case of brain metastasis. J Neurooncol. 2010;97(1):143–8.
Ashktorab H, Daremipouran M, Devaney J, Varma S, Rahi H, Lee E, et al. Identification of novel mutations by exome sequencing in African American colorectal cancer patients. Cancer. 2015;121(1):34–42.
Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/β-catenin signalling thresholds for tumourigenesis. Oncogene. 2013;32(39):4675–82.
Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23(2):228–35.
Gerecke C, Mascher C, Gottschalk U, Kleuser B, Scholtka B. Ultrasensitive detection of unknown colon cancer-initiating mutations using the example of the Adenomatous polyposis coli gene. Cancer Prev Res (Phila). 2013;6(9):898–907.
Rogler G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014;345(2):235–41.
Fox SA, Richards AK, Kusumah I, Perumal V, Bolitho EM, Mutsaers SE, et al. Expression profile and function of Wnt signaling mechanisms in malignant mesothelioma cells. Biochem Biophys Res Commun. 2013;440(1):82–7.
Valkenburg KC, Yu X, De Marzo AM, Spiering TJ, Matusik RJ, Williams BO. Activation of Wnt/β-catenin signaling in a subpopulation of murine prostate luminal epithelial cells induces high grade prostate intraepithelial neoplasia. Prostate. 2014;74(15):1506–20.
Zhang W, Jiao H, Zhang X, Zhao R, Wang F, He W, et al. Correlation between the expression of DNMT1, and GSTP1 and APC, and the methylation status of GSTP1 and APC in association with their clinical significance in prostate cancer. Mol Med Rep. 2015;12(1):141–6.
Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89.
Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8(9):e75885.
Jin G, Kim MJ, Jeon HS, Choi JE, Kim DS, Lee EB, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer. 2010;69(3):279–83.
Barnett CM, Heinrich MC, Lim J, Nelson D, Beadling C, Warrick A, et al. Genetic profiling to determine risk of relapse-free survival in high-risk localized prostate cancer. Clin Cancer Res. 2014;20(5):1306–12.
Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73(6):596–604.
Maire CL, Ligon KL. Molecular pathologic diagnosis of epidermal growth factor receptor. Neuro Oncol. 2014;16(Suppl 8):viii1–6.
Cho J, Bass AJ, Lawrence MS, Cibulskis K, Cho A, Lee SN, et al. Colon cancer-derived oncogenic EGFR G724S mutant identified by whole genome sequence analysis is dependent on asymmetric dimerization and sensitive to cetuximab. Mol Cancer. 2014;13:141.
Hour TC, Chung SD, Kang WY, Lin YC, Chuang SJ, Huang AM, et al. EGFR mediates docetaxel resistance in human castration-resistant prostate cancer through the Akt-dependent expression of ABCB1 (MDR1). Arch Toxicol. 2015;89(4):591–605.
National Center for Biotechnology Information USNLoM. ckit gene. http://www.ncbi.nlm.nih.gov/gene?term=(ckit%5bgene.
Database GTHG. kit gene. http://www.genecards.org/cgi-bin/carddisp.pl?gene=KIT&search=kit.
Simak R, Capodieci P, Cohen DW, Fair WR, Scher H, Melamed J, et al. Expression of c-kit and kit-ligand in benign and malignant prostatic tissues. Histol Histopathol. 2000;15(2):365–74.
Dakhova O, Rowley D, Ittmann M. Genes upregulated in prostate cancer reactive stroma promote prostate cancer progression in vivo. Clin Cancer Res. 2014;20(1):100–9.
Won D, Chi HS, Shim H, Jang S, Park CJ, Lee JH. The prognostic impact of c-KIT mutation in systemic mastocytosis associated with acute myeloid leukaemia patients. Leuk Res. 2013;37(8):883–8.
Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–54.
Segawa N, Nakamura M, Shan L, Utsunomiya H, Nakamura Y, Mori I, et al. Expression and somatic mutation on androgen receptor gene in prostate cancer. Int J Urol. 2002;9(10):545–53.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22.
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–86.
Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M. The androgen receptor gene mutations database: 2012 update. Hum Mutat. 2012;33(5):887–94.
Mononen N, Syrjäkoski K, Matikainen M, Tammela TL, Schleutker J, Kallioniemi OP, et al. Two percent of Finnish prostate cancer patients have a germ-line mutation in the hormone-binding domain of the androgen receptor gene. Cancer Res. 2000;60(22):6479–81.
Koochekpour S. Androgen receptor signaling and mutations in prostate cancer. Asian J Androl. 2010;12(5):639–57.
Li W, Cavasotto CN, Cardozo T, Ha S, Dang T, Taneja SS, et al. Androgen receptor mutations identified in prostate cancer and androgen insensitivity syndrome display aberrant ART-27 coactivator function. Mol Endocrinol. 2005;19(9):2273–82.
Zaman N, Giannopoulos PN, Chowdhury S, Bonneil E, Thibault P, Wang E, et al. Proteomic-coupled-network analysis of T877A-androgen receptor interactomes can predict clinical prostate cancer outcomes between White (non-Hispanic) and African-American groups. PLoS One. 2014;9(11):e113190.
Steinestel J, Luedeke M, Arndt A, Schnoeller TJ, Lennerz JK, Wurm C, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015. doi:10.18632/oncotarget.3925.
Jentzmik F, Azoitei A, Zengerling F, Damjanoski I, Cronauer MV. Androgen receptor aberrations in the era of abiraterone and enzalutamide. World J Urol. 2016;34(3):297–303.
Korpal M, Korn JM, Gao X, Rakiec DP, Ruddy DA, Doshi S, et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013;3(9):1030–43.
Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38.
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6(4):657–69.
Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69(1):16–22.
Steinkamp MP, O’Mahony OA, Brogley M, Rehman H, Lapensee EW, Dhanasekaran S, et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res. 2009;69(10):4434–42.
Ceder Y. Non-coding RNAs in prostate cancer: from discovery to clinical applications. Adv Exp Med Biol. 2016;886:155–70.
Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 2012;287(2):1527–37.
Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69(18):7165–9.
Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–64.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9.
Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121(7):626–33.
Zuhlke KA, Johnson AM, Tomlins SA, Palanisamy N, Carpten JD, Lange EM, et al. Identification of a novel germline SPOP mutation in a family with hereditary prostate cancer. Prostate. 2014;74(9):983–90.
Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346(6205):85–9.
Boysen G, Barbieri CE, Prandi D, Blattner M, Chae SS, Dahija A, et al. SPOP mutation leads to genomic instability in prostate cancer. Elife. 2015;4.
An J, Ren S, Murphy SJ, Dalangood S, Chang C, Pang X, et al. Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol Cell. 2015;59(6):904–16.
Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2016;35(4):403–14.
Robinson JL, Holmes KA, Carroll JS. FOXA1 mutations in hormone-dependent cancers. Front Oncol. 2013;3:20.
Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Grönberg H, et al. The mutational landscape of prostate cancer. Eur Urol. 2013;64(4):567–76.
St John J, Powell K, Conley-Lacomb MK, Chinni SR. TMPRSS2-ERG fusion gene expression in prostate tumor cells and its clinical and biological significance in prostate cancer progression. J Cancer Sci Ther. 2012;4(4):94–101.
García-Flores M, Casanova-Salas I, Rubio-Briones J, Calatrava A, Domínguez-Escrig J, Rubio L, et al. Clinico-pathological significance of the molecular alterations of the SPOP gene in prostate cancer. Eur J Cancer. 2014;50(17):2994–3002.
Ueda M, Iguchi T, Masuda T, Nakahara Y, Hirata H, Uchi R, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget. 2016. doi:10.18632/oncotarget.11409.
Mooney SM, Jolly MK, Levine H, Kulkarni P. Phenotypic plasticity in prostate cancer: role of intrinsically disordered proteins. Asian J Androl. 2016;18(5):704–10.
Beltran H, Antonarakis ES, Morris MJ, Attard G. emerging molecular biomarkers in advanced prostate cancer: translation to the clinic. Am Soc Clin Oncol Educ Book. 2016;35:131–41.
Wang S, Song Y, Yan F, Liu D. Mechanisms of resistance to third-generation EGFR tyrosine kinase inhibitors. Front Med. 2016. doi:10.1007/s11684-016-0488-1.
Pashayan N, Duffy SW, Chowdhury S, Dent T, Burton H, Neal DE, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104(10):1656–63.
Cybulski C, Wokołorczyk D, Kluźniak W, Kashyap A, Gołąb A, Słojewski M, et al. A personalised approach to prostate cancer screening based on genotyping of risk founder alleles. Br J Cancer. 2013;108(12):2601–9.
Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer. 2015;137(2):262–6.
Decker B, Karyadi DM, Davis BW, Karlins E, Tillmans LS, Stanford JL, et al. Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors. Am J Hum Genet. 2016;98(5):818–29.
Wojnarowicz PM, Oros KK, Quinn MC, Arcand SL, Gambaro K, Madore J, et al. The genomic landscape of TP53 and p53 annotated high grade ovarian serous carcinomas from a defined founder population associated with patient outcome. PLoS One. 2012;7(9):e45484.
Shanmugam V, Ramanathan RK, Lavender NA, Sinari S, Chadha M, Liang WS, et al. Whole genome sequencing reveals potential targets for therapy in patients with refractory KRAS mutated metastatic colorectal cancer. BMC Med Genomics. 2014;7:36.
Nakayama S, Sng N, Carretero J, Welner R, Hayashi Y, Yamamoto M, et al. β-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 2014;74(20):5891–902.
Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, et al. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics. 2014;7:32.
Muñoz-Moreno L, Arenas MI, Carmena MJ, Schally AV, Prieto JC, Bajo AM. Growth hormone-releasing hormone antagonists abolish the transactivation of human epidermal growth factor receptors in advanced prostate cancer models. Invest New Drugs. 2014;32(5):871–82.
Rapisuwon S, Parks K, Al-Refaie W, Atkins MB. Novel somatic KIT exon 8 mutation with dramatic response to imatinib in a patient with mucosal melanoma: a case report. Melanoma Res. 2014;24(5):509–11.
Gavert N, Shvab A, Sheffer M, Ben-Shmuel A, Haase G, Bakos E, et al. c-Kit is suppressed in human colon cancer tissue and contributes to L1-mediated metastasis. Cancer Res. 2013;73(18):5754–63.
Han X, Zhao J, Ji Y, Xu X, Lou W. Expression of CK19 and KIT in resectable pancreatic neuroendocrine tumors. Tumour Biol. 2013;34(5):2881–9.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
M. J. Alvarez-Cubero; L. J. Martinez-Gonzalez; I. Robles-Fernandez; J. Martinez-Herrera MS; G. Garcia-Rodriguez; M. Pascual-Geler MD; J. M. Cozar and J. A. Lorente, no have nothing to disclose. The authors have no conflict of interest.
Rights and permissions
About this article
Cite this article
Alvarez-Cubero, M.J., Martinez-Gonzalez, L.J., Robles-Fernandez, I. et al. Somatic Mutations in Prostate Cancer: Closer to Personalized Medicine. Mol Diagn Ther 21, 167–178 (2017). https://doi.org/10.1007/s40291-016-0248-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-016-0248-6