Abstract
The aim of the present study was to test the binding affinity of methylxanthines (caffeine/theine, methylxanthine, theobromine, theophylline and xanthine) to three potential target proteins namely Spike protein (6LZG), main protease (6LU7) and nucleocapsid protein N-terminal RNA binding domain (6M3M) of SARS-CoV-2. Proteins and ligand were generated using AutoDock 1.5.6 software. Binding affinity of methylxanthines with SARS-CoV-2 target proteins was determined using Autodock Vina. MD simulation of the best interacting complexes was performed using GROMACS 2018.3 (in duplicate) and Desmond program version 2.0 (academic version) (in triplicate) to study the stabile interaction of protein–ligand complexes. Among the selected methylxanthines, theophylline showed the best binding affinity with all the three targets of SARS-CoV-2 (6LZG − 5.7 kcal mol−1, 6LU7 − 6.5 kcal mol−1, 6M3M − 5.8 kcal mol−1). MD simulation results of 100 ns (in triplicate) showed that theophylline is stable in the binding pockets of all the selected SARS-CoV-2 proteins. Moreover, methylxanthines are safer and less toxic as shown by high LD50 value with Protox II software as compared to drug chloroquine. This research supports the use of methylxanthines as a SARS-CoV-2 inhibitor. It also lays the groundwork for future studies and could aid in the development of a treatment for SARS-CoV-2 and related viral infections.
Graphical abstract

Similar content being viewed by others
Introduction
SARS-CoV-2 is an enveloped; single stranded positive sense RNA virus which affects the respiratory tract of warm-blooded animals, including humans [1]. SARS-CoV-2 is associated with the common cold, pneumonia and severe acute respiratory syndrome (SARS). Coronavirus was first isolated in 1937 from an infectious bronchitis virus in chickens, which decimated the poultry population. These viruses cause 15–30% of all common colds (https://www.medicalnewstoday.com/articles/256521). Human coronavirus was discovered in the year 1960 in the nose of a patient suffering from common cold, and OC43 and 229E were the two human coronaviruses responsible for common cold [2]. A novel coronavirus, formally named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused coronavirus infection 2019 (COVID-19) around the world. According to World Health Organization (WHO), SARS-CoV-2 has caused a beginning outbreak in Wuhan, city of China [3••]. China was initially unaware of the direct transmission of the 2019 novel coronavirus (2019-nCoV). However, patients without symptoms were recognized as the major source of infection, and human-to-human transmission was also confirmed [4]. Four structural proteins have been identified in SARS-CoV-2, i.e. envelope, membrane, nucleocapsid and spike proteins [5]. The envelope and membrane proteins play an important role in the assembly of coronavirus. In contrast, nucleocapsid protein is needed for viral RNA synthesis [6]. In SARS-Cov-2, virus binding and its entry are governed by critical spike glycoprotein having subunits S1 with 685 amino acids and S2 with 588 amino acids [5, 7]. S1 subunit has receptor binding domain (RBD) that facilitates the entry of the virus into the host cells via ACE2 (angiotensin-converting enzyme 2) receptor [8]. External sub-domain of RBD has highest variation of amino acids, which accounts for direct interaction between the host ACE2 receptor and the spike protein of the virus [5, 8]. S2 subunit remains conserved among the SARS-CoV-2 viruses and shows 99% similarity with the bat SARS-CoVs [5]. The receptor binding domain (RBD) of SARS CoV-2 spike protein binds more strongly to human ACE2 receptors than SARS-CoV and can act as main target to prevent the viral attachment to ACE2-expressing cells [9]. The SARS-CoV-2 pandemic has taken a huge toll across the world, while therapeutics are still being discovered or developed. The primary target for most of the viral diseases including SARS-CoV-2 is the upper or lower respiratory tracts. Apart from vaccines, there are many natural phytocompounds that have a protective effect on the respiratory system. To build up the drug discovery pipeline for the treatment of SARS-CoV-2 that majorly affects the respiratory system, scientists across the world are taking new approaches such as repurposing the existing antiviral and anti-malarial drugs and in silico approach to explore and identify new molecules or phytocompounds that could be further tested under in vitro and in vivo systems. The in silico approach could help in building the repository and pipeline for clinical trials and reduce the time for discovering new drug candidates.
One such important class of compounds is methylxanthines present in coffee, tea and cacao [10], The most famous beverage and consumed by two-thirds of the world’s population. Caffeine/thiene, theobromine and theophylline are the most important methylxanthines.
Drinking beverages such as tea and coffee has been considered as health-promoting benefits since old-fashioned times. Caffeine-related methylxanthines are products of caffeine metabolism in vivo (paraxanthine, theobromine and theophylline). Theophylline is in clinical use for the treatment of asthma and is a well-known bronchodilator and reduces the symptoms of asthma, wheezing, coughing and breathlessness. Theophylline has also been reported as anti-inflammatory. The inhibition of phosphodiesterase (PDE) 3 is thought to cause bronchodilation, but the anti-inflammatory activity may be due to inhibition of PDE4 and activation of histone deacetylase-2, which switches off the activated inflammatory genes [11]. Theophylline reverses corticosteroid resistance via this process, which may be essential in extreme asthma and COPD (chronic inflammatory lung disease), where histone deacetylase-2 activity is reduced (Barnes, 2013). Caffeine (1,3,7-trimethylxanthine) is a methyl-xanthine alkaloid that is gaining popularity due to its wide range of physiological and pharmacological effects [12, 13]. Caffeine has been shown to have antiviral properties against HSV-1 (1,2). Yamazaki and Tagaya (1980) [14] published tentative findings of caffeine’s antiviral activity in viruses such as poliovirus, influenza virus, HSV-1 and vaccinia virus. Olson and Consigli (1979) [15] reported that caffeine has antiviral properties against NDV (Newcastle disease virus) by inhibiting the synthesis of viral RNA and proteins in infected cells. According to current consensus [12, 14,15,16,17], caffeine tends to prevent the development of NDV, human immunodeficiency virus, polyomavirus, HSV-1 and vaccinia virus. Caffeine and other methylxanthines have recently been shown to inhibit the replication of infectious HIV-1 strains, with the integration stage of the HIV-1 life cycle being the caffeine’s target [18]. Keeping in mind the importance of methylxanthines in inhibiting certain classes of viruses and health-promoting benefits, the current study was designed to test the interaction of methylxanthines (caffeine/thiene, methylxanthine, theobromine, theophylline, xanthine) with three different target proteins (spike, main protease and RNA protein) of SARS-CoV-2 using in silico tools (Fig. 1).
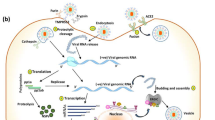
Hypothetical binding mechanisms of methylxanthines to SARS-CoV-2. Methylxanthines binds spike protein (6LZG), nucleocapsid (6M3M) and main protease (6LU7). Based on our in silico studies, it is hypothesized that methylxanthine competes with spike protein of SARS-CoV-2 for binding to ACE2 receptor. The proposed interactions between methylxanthines and ACE2 receptor need to be experimentally verified in the future. Normal entry of SARS-CoV-2 through ACE2 receptor is shown in the boxed part of the diagram
Methods
Bioinformatic Tools
Open Babel GUI [19], UCSF Chimera 1.8.1 [20]. Pubchem (www.pubchem.com), RCSB PDB (http://www.rscb.org/pdb), PDBsum (www.ebi.ac.uk/pdbsum), SwissADME and Autodock vina were used in the present investigation.
Ligand Preparation
Five major methylxanthines such as caffeine/thine, methylxanthine, theobromine, theophylline and xanthine (which are either found in coffee or the degradation products of caffeine) were selected in the current study. The 3-dimensional structures of all the methylxanthines, chloroquine (standard antimalarial drug) and lopinavir (standard antiretroviral drug) were retrieved from the pubchem (www.pubchem.com) in.sdf format. Open Babel was used to convert the phytocompounds.sdf file into PDB format [21]. Table 1 shows the molecular structure, compound CID and molecular weight of selected phytocompounds chloroquine (standard antimalarial drug) and lopinavir (standard antiretroviral drug).
Retrieval of SARS CoV-2 Proteins
The crystal structures of three SARS CoV-2 proteins were downloaded in pdb format from the protein databank, namely nucleocapsid protein N-terminal RNA binding domain (PDB ID: 6M3M) [22], main protease (6LU7) [23] and spike receptor-binding domain (PDB ID: 6LZG) [24]. For virtual screening, xanthine derivatives such as caffeine/thiene, methylxanthine, theobromine, theophylline and xanthine were used.
Target Protein Preparation
The high-resolution structure of SARS-CoV2 spike receptor-binding domain (PDB ID: 6LZG as shown in Fig. 2A,B), main protease (PDB ID: 6LU7 shown in Fig. 2C,D), nucleocapsid protein N-terminal RNA binding domain (PDB ID: 6M3M shown in Fig. 2E,F) and the Auto-Dock tool 1.5.4 was used for the preparation of the protein structures. The selected protein structures were processed for creating disulfide bonds, assigning proper bond orders and the addition of missing hydrogens in the raw structure. Hydrogen bond optimization was assigned using non-hydrogen atoms of protein.
Structures of selected SARS-CoV-2 S proteins: spike protein 6LZG (A: surface view representation and B: secondary structure representation); main protease 6LU7 (C: surface view representation and D: secondary structure representation); nucleocapsid protein N-terminal RNA binding domain (E: surface view representation and F: secondary structure representation). Helices are shown in red, -sheets in yellow and loops in green color (A–F)
ADMET and Toxicity Prediction of Phytocompounds
Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) screening was done to determine the absorption, toxicity and drug-likeness properties of selected methylxanthines (ligands). The 3D structures of ligands (caffeine, methylxanthine, theobromine, theophylline, thiene and xanthine) were saved in.smiles format and were uploaded on SWISSADME (Molecular Modeling Group of the SIB (Swiss Institute of Bioinformatics), Lausanne, Switzerland), admetSAR (Laboratory of Molecular Modeling and Design, Shanghai, China) and PROTOX webservers (Charite University of Medicine, Institute for Physiology, Structural Bioinformatics Group, Berlin, Germany) for ADMET screening. SWISSADME is a web tool used for the prediction of ADME and pharmacokinetic properties of a molecule. The predicted outcome includes lipophilicity, water solubility, physicochemical properties, pharmacokinetics, drug resemblance, medicinal chemistry and the Brain or Intestinal Estimated Permeation Method (blood–brain barrier and PGP prediction) [25]. admetSAR can predict about fifty ADMET properties and provides ADMET profiles for the query molecules. The following are the toxicity indicators: classes: I Compounds with LD50 values less than 50 mg kg−1, (ii) Compounds with LD50 values greater than 50 mg kg−1 but less than 500 mg kg−1 (Cheng et al., 2012; Yang et al., 2019). PROTOX is a server that predicts the LD50 value and toxicity class of a question molecule in rodents. The following are the toxicity classes: (Class 1: fatal if swallowed (LD50 5) and Class 2: fatal if swallowed (55,000) [26, 27••, 28••].
Molecular Docking
All the xanthine derivatives were used to perform molecular docking studies against three different proteins of SARS-CoV-2 to find the potential hits for further drug discovery experiments. In the current study, AutoDock vina was used for performing the docking studies [29, 30]. AutoDock vina uses a Lamarckian Genetic Algorithm (LGA) and is based on a semi empirical free energy force field. In all the proteins, the grid was manually adjusted within the key active binding amino acid residues. The high scoring compounds were chosen based on the binding energy of the ligands with the receptor. The lower ligand binding energy with the receptor is an indication of high affinity for the target receptor.
The.pdb complex of protein and ligands was analyzed by discovery studio (www.ebi.ac.uk/pdbsum) to classify the protein–ligand interactions. Chimera performed detailed visualization and comparison of docked sites of target proteins and ligands.
Molecular Dynamics Simulation
MD simulation was done to study the stability of protein ligand complexes over the period of 100 ns. For the authenticity of the data, MD simulation was done with two different software such as GROMACS 2018.3 (simulation in duplicate) and Desmond program version 2.0 academic version (simulation in triplicate). Among all the selected compounds, theophylline showed the best interactions with three selected targets (6LZG1, 6LU7 and 6M3M) of SARS-CoV-2.
MD Simulation by Using GROMACS 2018.3 Software
Molecular dynamics study of the theophylline complexes with 6LZG1, 6LU7 and 6M3M was done using GROMACS 2018.3 in duplicate [31] software which was installed in ubuntu 18.04 LTS. Docked structures of the protein–ligand complexes (theophylline with 6LZG1, 6LU7 and 6M3M) were used in the simulation study. Proteins were processed, and the topology file was prepared by using pdb2gmx and GROMOS54a7_atb.ff (imported from the automated topology builder website) force field, while the ligand topology file was prepared by using the Automated Topology Builder (ATB) version 3.0. The solvent addition was done in a cubic box by using a box distance of 1.0 nm from closest atom in the protein. To neutralize the device, the Cl− ions calculated from the genion module for each protein were used. The energy was minimized by using the steepest descent algorithm with 50,000 steps and a cumulative force of 5 kJ mol−1, as well as the Verlet cutoff scheme with particle mesh Ewald (PME) columbic interactions. During the equilibration process, position restraints were used. Following that, NVT equilibration was performed at 300 K with 100 ps steps, and NPT equilibration was performed with Parrinello-Rahman (pressure coupling), 1 bar reference pressure and 100 ps steps. The LINCS algorithm was used to constrain the length of all bonds. For long-range electrostatics, the particle mesh Ewald (PME) algorithm was used. The protein–ligand complex’s MD was run for 100 ns (in duplicate). Following efficient completion of a 100-ns molecular dynamic simulation, the root means square deviation (RMSD) of backbone residues, the number of hydrogen bonds and the root mean square fluctuations (RMSF), radius of gyration (RG) and solvent accessible surface area (SASA) were calculated.
Estimation of Free Energy of Binding
The free energy of binding calculations was done using the standalone program, g mmpbsa, following the protocol described by Kumari et al. (2014) [32]. Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) is a widely used method to calculate the free binding energy and to predict the stability of the complex. Modified molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) is an open-source software and employed to calculate the free energy of binding between two defined groups. Recently, MM-PBSA algorithm has been utilized as a scoring function in computational drug design [33, 34]. The free energy of binding is based on the theory:
where Gcomplex is the total free energy of the protein-inhibitor complex, and Gprotein and Gligand are total free energies of the separated form of protein and inhibitor in solvent, respectively [33]. The average binding energy calculations were done by a python script provided in g mmpbsa stand-alone program.
MD Simulation by Using Desmond Program Version 2.0 (Academic Version)
To check the accuracy of docking observations and the stability of protein–ligand interactions, dynamic simulations (MD) simulations were performed in triplicate using the Desmond software version 2.0 (academic version) [28••, 30, 34,35,36,37,38]. All the three complexes were solvated in TIP3P water model with cubic periodic box containing Simple Point Charge (SPC) (101,010) with Optimized Potentials for Liquid Simulations (OPLS) all-atom force field 2005. MD simulation was performed to check the accuracy of docking observations and the stability of protein–ligand interactions. To run the MD simulations, the OPLS 2005 force field parameters were combined with periodic boundary conditions in the NPT ensemble method, which included a relaxation time of 1 ps at a constant temperature of 300 K, a constant length, a smooth particle mesh Ewald (PME) (with a 10–9 tolerance limit) and a cutoff distance of 9.0 Å. 100-ns production time had been chosen for the analysis of protein structure evaluation by every 1 ns. To decipher the stability, an average structure was chosen from the MD simulation corresponding to production phase. In addition, root mean square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bond, radius of gyration (Rg), histogram for torsional bonds were performed for the investigation of structural changes with dynamic role of the protein–ligand complexes for 100 ns. After successful completion of the MD simulation for 100 ns, MM-GBSA calculation was done in triplicates.
Results
ADMET Prediction and Toxicity Analysis of Selected Phytocompounds
The ADMET properties predicted by SwissADME of selected phytocompounds and drugs Chloroquine and Lopinavir are summarized in Table 2. A Log Po/w value of 5 implies strong aqueous solubility, which means that a sufficient amount of drug will enter and stay within the body after oral administration. The Log Po/w value of all the selected phytocompounds is 5 (Table 2). The capacity of a compound to permeate into the cells is indicated by TPSA (total polar surface area). For good permeation of the compound through the cell membrane, a TPSA value of 94.40 Å is needed, and a value of 90 Å is required to pass through the blood–brain barrier. Except for xanthine, all other phytocompounds (caffeine/thiene, methylxanthine, theobromine and theophylline) showed a TPSA value of 90 Å, indicating good permeability through blood–brain barrier. Lipinski’s rule of five aids in determining a compound’s drug likeness; an orally active drug does not break more than one of Lipinski’s rules. None of the selected ligands violated any of Lipinski’s rule of five (Table 2). Table 3 summarizes the toxicity expected by PROTOXII and admetSAR. Table 3 shows that all the selected phytocompounds are non-carcinogenic and non-cytotoxic in nature and are thus safe to use. Caffeine/thiene, methylxanthine, theobromine, theophylline and xanthine had lower LD50 values than all other phytocompounds as measured using Protox II.
Docking Analysis
The selected phytocompounds showed relatively good binding affinity with all the selected target proteins of SARS CoV-2 (spike receptor-binding domain PDB ID: 6LZG, nucleocapsid protein N-terminal RNA binding domain PDB ID: 6M3M and main protease PDB ID: 6LU7) of SARS-CoV-2. Theophylline showed the best interaction binding affinity with 6LZG (− 5.7 kcal mol−1), followed by methylxanthine (-5.3 kcal mol−1), xanthine (− 5.2 kcal mol−1), theobromine (− 4.9 kcal mol−1) and caffeine (− 4.8 kcal mol−1) as shown in Figs. 3, 4 and Table 4. Interestingly, nucleocapsid protein N-terminal RNA binding domain (6M3M) also showed best interaction with theophylline (− 5.8 kcal mol−1), followed by theobromine (− 5.5 kcal mol−1), xanthine (− 4.9 kcal mol−1), methylxanthine (− 4.7 kcal mol−1) and caffeine (− 4.6 kcal mol−1) as shown in Figs. 4, 5, 6 and Table 4. Similarly, the main protease (6LU7) also showed best binding interaction with theophylline (kcal mol−1 − 6.5), followed by theobromine (− 5.8 kcal mol−1), caffeine (− 5.7 kcal mol−1), methylxanthine and xanthine (− 5.5 kcal mol−1) as shown in Figs. 7, 8 and Table 4. Binding affinity of Lopinavir was much stronger (6LZG: − 7.1 kcal mol−1, 6M3M: − 6.4 kcal mol−1, 6LU7: − 8.2 kcal mol−1) as compared to selected phytocompounds (Table 4) and chloroquine also represented in Table 4. In contrast to Lopinavir, Chloroquine showed weak interaction with all the three targets (6LZG: − 5.1 kcal mol−1, 6M3M: − 4.9 kcal mol−1, 6LU7: − 5.7 kcal mol−1) but comparable to the selected compounds (Table 4). The results of this study suggested that theophylline showed the best interaction with all the three selected target protein of SARS-CoV-2. Complexes of theophylline with 6LZG, 6M3M and 6LU7 were further analyzed by MD simulation.
Electrostatic surface view representation of receptor-binding domain (PDB ID: 6LZG) with bound xanthine derivatives: (A) caffeine, (B) chloroquine, (C) methylxanthine, (D) theobromine, (E) theophylline and (F) xanthine. The color coding is by electrostatic potential; negative, positive and neutral regions are shown in blue, red and white respectively (image generated using Pymol)
Electrostatic surface view representation of nucleocapsid protein N-terminal RNA binding domain (PDB ID: 6M3M) with bound xanthine derivatives: (A) caffeine, (B) chloroquine, (C) methylxanthine, (D) theobromine, (E) theophylline and (F) xanthine. The color coding is by electrostatic potential; negative, positive and neutral regions are shown in blue; red and white respectively (image generated using Pymol)
Electrostatic surface view representation of SARS-CoV-2 main protease (PDB ID: 6LU7) with bound xanthine derivatives: (A) caffeine, (B) chloroquine, (C) methylxanthine, (D) theobromine, (E) theophylline and (F) xanthine. The color coding is by electrostatic potential; negative, positive and neutral regions are shown in blue, red and white respectively (image generated using Pymol)
Each amino acid residue is numbered followed by three letter codes. Amino acid residues in boldface letters are the phosphorylation sites on serine and threonine, predicted by Davidson et al. (2020).
MD Simulation by Using GROMACS 2018.3 Software
MD simulation of theophylline in complex with 6LZG, 6LU7, 6M3M was done for validation of molecules docking study and stability of theophylline in complex with SARAS-CoV-2 proteins (6LZG, 6LU7 and 6M3M). MD simulation was done for 100 ns.
RMSD Analysis of Protein–Ligand Complexes
RMSD of theophylline in complex with 6LZG was stabilized at 10ns/0.4 nm in both the runs. RMSD of theophylline in complex with 6LZG remains stable up to 80ns and further stabilized at 90–100ns at 0.7–0.8 nm in first run. No major fluctuation was observed in the second run (Fig. 9A), and minor fluctuations are however still within the acceptable limits. The RMSD of theophylline in complex with 6LU7 and 6M3M was stable throughout the entire simulation (Figure 9B,C) during both the runs. The estimated RMSD for the complexes shows that the theophylline complex with 6LU7 and 6M3M is more stable as compared to the spike protein (6LZG).
RMSF Analysis of Protein–Ligand Complexes
RMSF was done to study the flexibility of protein-ligand complexes and fluctuation in interactive amino acid residues in secondary structure of target proteins (6LZG, 6LU7 and 6M3M) SARS-CoV-2.
The ligand RMSF plot for theophylline fit over the entire 6LZG protein and showed very minor residual fluctuation in alpha helical and beta strands. The fluctuation was observed in ARG 127 and TYR 143 during both the runs (Fig. 10A). In case of 6LU7 complex with theophylline, no residual fluctuation was observed in alpha helical and beta strands in both runs (Fig. 10B). Similarly, in case of 6M3M with theophylline, we observed a perfect fit within the binding pocket of target protein, and residual fluctuation was observed at ALA 91 in both the runs of simulation (Fig. 10C).
Radius of gyration is in the range of 1.8–1.95 nm for 6LZG complex with theophylline, 2.2–2.5 nm for 6LU7 in complex with theophylline and 1.35–1.4 nm for 6M3M in complex with theophylline during both the runs as shown in Figure 11A, B, C. The radius of gyration plots establishes the compactness of the theophylline complexes with the SARS-CoV-2 proteins (6LZG, 6LU7 and 6M3M) and confirms their stable interactions. Solvent accessible range of theophylline complex with 6LZG is between 106 and 125nm, theophylline complex with 6LU7 is between 145 and 160 nm and theophylline complex with 6M3M is 73–84 nm during both the runs as shown in supplementary figures (Fig. S1A, B, C). The changes in the solvent accessible surface area did not change much about a mean position through the course of the simulation of theophylline with the SARS-CoV-2 proteins (6LZG, 6LU7 and 6M3M). This suggest that the interaction of the ligands with the selected proteins did not overtly change the conformation of the protein about the mean position. However, the hydrogen bonding (Fig. S2A, B, C) observed for the three complexes during the course of the simulation showed that theophylline had the most polar interactions with the 6LU7 protein, which must have ultimately contributed to it having the highest estimated free energy of binding. Essentially, the data obtained from the RMSD, RMSF, ROG, SASA and hydrogen bonding estimates are in good agreement with the observed free energy of binding for the three complexes of theophylline with the SARS-CoV-2 target proteins (6LZG, 6LU7 and 6M3M). The complexes are stable, and the changes over the course of the simulation are limited, but significantly different from each other. The free energies are different, and the highest free energy of binding was observed for the theophylline complex with 6LU7. The breakdown of the contributions of all the energy terms to the binding energy is presented in Table 5.
Radius of gyration graph of best protein–ligand complexes of theophylline with SARS-CoV-2 proteins for 100 ns: (A) theophylline complex with 6LZG, (B) theophylline complex with 6LU7 and (C) theophylline complex with 6M3M. The duplicate runs are indicated by different line colors as indicated in A, B and C panels
MD Simulation by Using Desmond Program Version 2.0 (Academic Version)
MD simulation of theophylline in complex with 6LZG, 6LU7 and 6M3M was done in triplicate (100ns) to study the stability of protein-ligand complexes using Desmond program version 2.0 to verify the binding stability.
RMSD of Protein–Ligand Complexes
The RMSD graph of theophylline in complex 6LZG showed the stability of backbone and Cα at 40 ns at 4–4.5 Å and remains stable up to 100-ns time scale. The RMSD of ligand seems to be stable from the beginning of simulation (0.2–0.5 Å) and remains stable up to 100 ns as shown in Fig. 12A. RMSD of theophylline in complex with 6LU7 showed stability in the backbone and Cα at 30 ns at 3.2–3.6 Å. Ligand RMSD was stable from the beginning of simulation as shown in Fig. 12B. In case of theophylline complex with 6LZG, RMSD of backbone (3–3.5 Å) and Cα (2–2.5 Å) was stable from the initial phase of simulation and remains stable up to 100 ns. RMSD of the ligand was stable from 0 to 100 ns at 0.5 Å (Fig. 12C).
RMSD graph of best protein–ligand complexes of theophylline with SARS-CoV-2 proteins for 100 ns: (A) theophylline complex with 6LZG, (B) theophylline complex with 6LU7 and (C) theophylline complex with 6M3M. The different color lines for Cα, backbone, side chains, heavy atoms, ligand fit on protein and ligand fit on ligand are as indicated at the top of panel A, B and C
RMSF of Protein–Ligand Complexes
RMSF explained the flexibility of protein structures and fluctuation of interactive residues in secondary structure elements of target proteins. The RMSF plot for theophylline in complex with 6LZG showed low residual fluctuation in α-helical and β-strands (Fig. 13A) and showed hydrophobic interactions with Val 73, Ile 75; hydrogen bonds with Gln 71, Pro 75, Asn 76 and water bridges with Gly 70, Gln 71, Gln 84, Tyr 113. (Fig. 14A). RMSF plot for theophylline fits over the entire 6LU7 protein, and no residual fluctuation was observed in α-helical and β-strands (Fig. 13B). Theophylline in complex with 6LU7 showed hydrophobic interactions with Met 49, Met 165, Leu 167, Ala 173, Phe 185; hydrogen bond interactions with His 41, Cys 44, Asn 142, Gly 143, Cys 145, His 164, Arg 188, Gln 189, Thr 190 and water bridges with Thr 26, Arg 40, His 41, Asn 119, His 164, Glu 166, Val 185, Val 186, Asp 187, Arg 188, Gln 189, Thr 190, Gln 192, Ala 193 (Fig. 14B). Similarly, RMSF of theophylline fit over the entire 6M3M, and no residual fluctuation was observed in α-helical and β-strands (Fig. 14C). Theophylline in complex with 6M3M showed the hydrophobic interactions with Phe12, Leu 122, Leu 125, Phe 126, Lys 128, Tyr 143, Phe 156, Tyr 159, Phe 160, Leu 162, Leu 188; hydrogen bonding with Ser 41, Tyr 91, Arg 124, Lys 128, Ser 129, Glu 129, Glu 141, Ser 147, Asn 151, Glu 154, Gly 155, Phe 156, Lys 156, Asn 157, Lys 158, Tyr 159, Phe 160, Leu 187, His 189 and water bridges with Asn 40, Lys 87, Tyr 91, Arg 124, Leu 125, Arg 127, Lys 128, Ile138, Ser 139, Glu 141, Tyr 143, Gln 144, Ala 445, Thr 148, Pro 149, Asn 157, Lys 158, Tyr 159, Phe 160, Leu 162, Gln 163, Ser 164, Glu 186, Leu 187, His 189 (Fig. 14C). The breakdown of the contributions of all the energy terms to the binding energy is presented in Table 6.
RMSF graph of best protein–ligand complexes of theophylline with SARS-CoV-2 proteins for 100 ns: (A) theophylline complex with 6LZG, (B) theophylline complex with 6LU7 and (C) theophylline complex with 6M3M. The different color lines for Cα, backbone, side chains, heavy atoms and B factor are as indicated at the top of panel A, B and C
Histogram of ligand contacts with amino acid residues of target proteins: (A) theophylline complex with 6LZG, (B) theophylline complex with 6LU7 and (C) theophylline complex with 6M3M. The different colors indicating H-bonds, hydrophobic, ionic and water bridges are as indicated at the bottom of panel C
Discussion
Molecular docking and computational techniques are widely used to predict drug likeness, toxicity and protein–ligand interactions for speeding up drug discovery. Molecular docking and in silico approach could help in building the pipeline for wet lab experiments and clinical trials and advance the drug discovery challenges. The current study was focused on major xanthine derivative phytocompounds present in coffee and tea to provide relief and reduce the symptoms of Covid-19 infection and related viruses. The research idea stems from the fact that theophylline (a known methylxanthine) acts as a bronchodilator by relaxing the muscles in the lungs and widening the airways (bronchi). Nasal and lungs are the primary target expressing ACE2 receptor and spike protein of SARS-CoV-2, and related viruses bind ACE2 receptor and initiate pathogenesis.
Molecular docking approach is one technique that helps in identification of interacting molecules. Lower score in molecular docking is considered as higher stability in ligand receptor binding [39]. Using molecular docking approach, we found that caffeine, methylxanthine, theobromine, theophylline, thiene and xanthine are effective in binding affinity or pharmacokinetic properties with three different proteins (6LZG, 6LU7 and 6M3M) of SARS-CoV-2. Among all the selected phytocompounds, theophylline showed most promising interaction with all the three different targets of SARS-CoV-2 with docking scores of − 5.7, − 6.5 and − 5.8 kcal mol−1 with 6LZG, 6LU7 and 6M3M respectively. Similar to our study, Celik et al. (2020) [40••] reported the mechanism of hydroxychloroquine on SARS-CoV-2 proteins with docking scores − 3.074 (6VXX) and − 6.227 (6LZG). Peele et al. (2020) [41] screened 68 US-FDA approved drugs and phytocompounds for binding with SARS-CoV-2 main protease (6LU7), and the best docking score was found with lopinavir, amodiaquine and theaflavin digallate. Bhowmik et al. (2020) reported the antiviral activity of rutin, doxycycline, caffeic acid, ferulic acid, simeprevir and grazoprevir with envelop (E), membrane (M), nucleocapsid (N) proteins of SARS-CoV-2 proteins. They found that rutin and doxycycline showed promising interaction with envelop protein (E) with binding scores of − 9.3 kcal mol−1 and − 9.1 kcal mol−1) respectively. On the other hand, caffeic acid and ferulic acid showed best binding with membrane protein (M)with binding scores of − 8.4 kcal mol−1 and − 8.3 kcal mol−1 respectively. Simeprevir and grazoprevir showed promising binding with nucleocapsid (N) protein with binding scores of − 8.7 kcal mol−1 and − 8.5 kcal mol−1 respectively. Utsunomiya et al. (2008) [42] reported the antiviral activity of hot water extract and instant coffee solution against coffee virus and herpes type 1 virus. Clarck et al. (1998) [43] reported the antiviral activity of crude theaflavin extracted from black tea against coronavirus with EC50 value of 34.7 µg ml−1. Caffeine inhibits the SARS coronavirus main peptidase (6LU7) [44••] and Paine like protease (6W9C) [45]. Theophylline is used as a bronchodilator, and at lower concentration it also showed anti-inflammatory effect in asthma and COPD [9]. Murayama et al. (2008) [46] reported that HSV1, poliovirus and influenza are sensitive to caffeine and have potential to inhibit both DNA and RNA viruses. Theophylline and pyrimidine derivatives are possible inhibitors of RNA bound N-terminal domain [47]. Weber et al. (2020) [48] reported tea infusions inhibited both adenovirus infection and viral protein adenain in HepG2 cells. A report from Clarck et al. (1998) [43] showed the anti-rotaviral activity of black tea theaflavin, whereas the crude tea extract was active against coronavirus. The black tea theaflavin also inhibited the replication of HIV [49]. Polyphenolic compounds of black tea inhibited HIV-1 entry into the target cell by blocking the HIV entry through its envelop (glycoprotein-mediated membrane fusion). Elzupis [50••] studied the inhibitory effect of caffeine and caffeine containing pharmaceuticals known as 3CPs (theophylline, linagliptin, dyphylline, bromotheophylline, pentoxifylline and istradefylline) against SARS-CoV-2 (3-chymotrypsin-like protease 3CLpro). This in silico study showed for the first time that inexpensive drugs available in large quantities could potentially act as inhibitors against 3CLpro. It was also suggested that linagliptin and caffeine could be more specifically used for the treatment of SARS-CoV-2 after in vitro and in vivo studies. LeBlanc and Colpitts [51] proposed that green tea catechin, epigallocatechin gallate (EGCG), prevents murine and human coronavirus infection. Jang et al. [52] showed that tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. EGCG and theaflavin (active ingredients of green tea and black tea) showed inhibitory activity against the SARS-CoV-2 3CL-protease with half inhibitory concentration (IC50) was 7.58 μg/ml for EGCG and 8.44 μg/ml for theaflavin. No cytotoxicity was observed for either EGCG or theaflavin at the concentrations tested up to 40 μg/ml in HEK293T cells. Bhardwaj et al. [53••] showed that Oolonghomobis flavan, a bioactive molecule found in tea, acts as an inhibitor for the Mpro of SARS-CoV-2. In spite of the tremendous health benefits and potential drug for treating various viral infections, Gottwalt and Tadi [54] have proposed that methylxanthines have a narrow therapeutic range. Mild effects such as nausea, vomiting, increased gastric acid secretion, polyuria, insomnia, palpitations, headaches and tremors could occur even below concentrations of 20 mcg/ml. Severe side effects such as intractable vomiting, arrhythmias, irregular heartbeat (slow or fast), cardiac arrest, allergic skin reactions or seizures could occur with serum concentrations that exceed 20 mcg/ml. Furge and Guengerich [55] have reported Cytochrome P450 may deactivate the caffeine and could reduce its efficacy.
Conclusion
Caffeine/thiene, methylxanthine, theobromine, theophylline and xanthine were screened on the basis of binding energy against SARS-CoV-2 spike protein S1 and receptor-binding domain (S1RBD). All the selected phytocompounds showed promising binding affinity with SARS-CoV-2 spike protein and S1 receptor-binding domain (S1RBD) and could potentially prevent the binding of virus to ACE2 receptor and stop the infection. ADMET prediction revealed that caffeine/thiene, methylxanthine, theobromine, theophylline and xanthine could be developed as potential drugs for SARS-CoV-2 treatment. Based on our in silico studies, the future challenge is to test all the methylxanthine derivatives using in vitro and in vivo assays for developing broad spectrum therapeutics against different viruses including SARS-CoV-2.
Availability of Data and Material
Supporting data is provided in supplementary data.
References
Papers of particular interest, published recently, have been highlighted as:•• of major importance
Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): An update. Cureus. 2020;e7423. https://doi.org/10.7759/cureus.7423
Abdul-Rasool S, Fielding BC. Understanding human coronavirus HCoV-NL63. Open Virol J. 2010;4:76. https://doi.org/10.2174/1874357901004010076
••Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dynam. 2020;1–6. https://doi.org/10.1080/07391102.2020.1752802. This in silico study explored the effect of synergism of the drugs, lopinavir, oseltamivir and ritonavir against the SARS-CoV-2 protease.
Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–1. https://doi.org/10.1056/NEJMc2001468.
Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infec. 2020;9:221–36. https://doi.org/10.1080/22221751.2020.1719902.
Schoeman D, Fielding BC. Corona virus envelope protein: current knowledge. Virol J. 2019;16:69. https://doi.org/10.1186/s12985-019-1182-0.
Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. https://doi.org/10.3390/v12020135.
Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virology. 2020;94:e00127-e220. https://doi.org/10.1128/JVI.00127-20.
Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–20. https://doi.org/10.1038/s41423-020-0400-4.
Bonetti F, Brombo G, Zuliani G. Nootropics, functional foods, and dietary patterns for prevention of cognitive decline. In Nutrition and Functional Foods for Healthy Aging. Academic Press. 2017;211–232. https://doi.org/10.1016/B978-0-12-805376-8.00019-8
Barnes PJ. Theophylline. Am J Respir Crit Care Med. American journal of respiratory and critical care medicine. 2013;901–906. https://doi.org/10.1164/rccm.201302-0388PP
Cowan MM. Plant products as antimicrobial agents. Clin Microbiol Rev. 1999;12:564–82. https://doi.org/10.1128/CMR.12.4.564.
Daly JW. Caffeine analogs: biomedical impacts. Cell Mol Life Sci. 2005;64:2153–69. https://doi.org/10.1007/s00018-007-7051-9.
Yamazaki Z, Tagaya I. Antiviral effect of atropine and caffeine. J Gen Virol. 1980;50:429–31. https://doi.org/10.1099/0022-1317-50-2-429.
Olson NJ, Consigli RA. Production of labile Newcastle disease virus progeny after infection of chicken embryo cells in the presence of caffeine. Ameri J Vet Res. 1979;40:387–92.
Daniel R, Marusich E, Argyris E, Zhao RY, Skalka AM, Pomerantz RJ. Caffeine inhibits human immunodeficiency virus type 1 transduction of nondividing cells. J Virol. 2005;76:2058–65. https://doi.org/10.1128/JVI.79.4.2058-2065.2005.
Dahl J, You J, Benjamin TL. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol. 2005;79:13007–17. https://doi.org/10.1128/JVI.79.20.13007-13017.2005.
Nunnari G, Argyris E, Fang J, Mehlman KE, Pomerantz RJ, Daniel R. Inhibition of HIV-1 replication by caffeine and caffeine-related methylxanthines. Virol. 2005;335:177–84. https://doi.org/10.1016/j.virol.2005.02.015.
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Chem Informat. 2011;3:33. https://doi.org/10.1186/1758-2946-3-33.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Computat Chem. 2004;25:1605–12.
Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, Gu M. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci. 2009;122:2055–63. https://doi.org/10.1242/jcs.049080.
Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q, Yan Y. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmace Sinica B. 2020;10:1228–38. https://doi.org/10.1016/j.apsb.2020.04.009.
Liu S, Lu H, Zhao Q, He Y, Niu J, Debnath AK, Wu S, Jiang S. Theaflavin derivatives in black tea and catechin derivatives in green tea inhibit HIV-1 entry by targeting gp41. Biochim Biophys Acta 2005; 1723(1-3):270–81. https://doi.org/10.1016/j.bbagen.2005.02.012
Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894-904.e9.
Salaria D, Rolta R, Sharma N, Dev K, Sourirajan A, Kumar V. In silico and in vitro evaluation of the anti-inflammatory and antioxidant potential of Cymbopogon citratus from North-western Himalayas.. bioRxiv. 2020, https://doi.org/10.1101/2020.05.31.124982
Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucl Acids Res. 2018;46:W257–63. https://doi.org/10.1093/nar/gky318.
••Rolta R, Salaria D, Kumar V, Sourirajan A, Dev K. Phytocompounds of Rheum emodi, Thymus serpyllum and Artemisia annua inhibit COVID-19 binding to ACE2 receptor: in silico approach. Curr Pharmacol Rep. 2021. https://doi.org/10.1007/s40495-021-00259-4. This In silico study showed that artemisinin stably interact with SARS-CoV-2 spike glycoproteins (6VXX and 6VYB), hence proposed as a potential phytochemical to prevent SARS-CoV-2 interaction with ACE-2 receptor.
••Rolta R, Yadav R, Salaria D, Trivedi S, Imran M, Sourirajan A, Baumler DJ, Dev K. In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J Biomol Struct Dynam. 2020;1–8. https://doi.org/10.1080/07391102.2020.1804457. This in silico study emodin, aloe-emodin, anthrarufin, alizarine, and dantron showed good binding affinity at all the three active sites of RNA binding domain of nucleocapsid phosphoprotein of COVID-19. All the active phytocompounds follows the drug likeness properties, non-carcinogenic, and non-toxic.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. https://doi.org/10.1002/jcc.21334.
Rolta R, Salaria D, Kumar V, Patel CN, Sourirajan A, Baumler DJ, Dev K. Molecular Docking studies of Phytocompounds of Rheum emodi Wall with proteins responsible for antibiotic resistance in bacterial and fungal pathogens: in silico approach to enhance the bio-availability of antibiotics. J Biomol Struct Dynam. 2020;1–15. https://doi.org/10.1080/07391102.2020.1850364
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;2:19–25. https://doi.org/10.1016/j.softx.2015.06.001.
Kumari R, Kumar R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Info Model. 2014;54:1951–1962. doi:https://doi.org/10.1021/ci500020m
Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem. 2000;43:3867–77. https://doi.org/10.1021/jm000292e.
Pant R, Joshi A, Maiti P, Nand M, Pande V, Chandra S. Identification of potential mycolyltransferase Ag85C inhibitors of Mycobacterium tuberculosis H37Rv via virtual high throughput screening and binding free energy studies. J Mol Graph Mod. 2020;98: 107584. https://doi.org/10.1016/j.jmgm.2020.107584.
Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD Salmon JK. Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 2006; 43–43. https://doi.org/10.1109/SC.2006.54.
Patel CN, Georrge JJ, Modi KM, Narechania MB, Patel DP, Gonzalez FJ, Pandya HA. Pharmacophore-based virtual screening of catechol-o-methyltransferase (COMT) inhibitors to combat Alzheimer’s disease. J Biomol Struct Dynam. 2018;36:3938–57. https://doi.org/10.1080/07391102.2017.1404931.
Patel CN, Kumar SP, Modi KM, Soni MN, Modi NR, Pandya HA. Cardiotonic steroids as potential Na+/K+-ATPase inhibitors—a computational study. J Recept Signal Transduct. 2019;39:226–34. https://doi.org/10.1080/10799893.2019.1660893.
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput. 2010;6:1509–19. https://doi.org/10.1021/ct900587b.
Krovat EM, Steindl T, Langer T. Recent advances in docking and scoring. Curr Comp Aided Drug Design. 2005;1:93–102. https://doi.org/10.2174/1573409052952314.
••Celı̇k I, Onay-Besı̇kcı̇ A, Ayhan-Kilcigı̇l G. Approach to the mechanism of action of hydroxychloroquine on SARS-CoV-2: a molecular docking study. J Biomol Struct Dynam. 2020;1–7. https://doi.org/10.1080/07391102.2020.1835721.The molecular docking study showedt that hydroxychloroquine and chloroquine prevent the binding of SARS-CoV-2 spike protein to ACE2 by interacting with the allosteric site.
Peele KA, Durthi CP, Srihansa T, Krupanidhi S, Ayyagari VS, Babu DJ, Indira M, Reddy AR, Venkateswarulu TC. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Info Medicine Unlock. 2020;19: 100345. https://doi.org/10.1016/j.imu.2020.100345.
Utsunomiya H, Ichinose M, Uozaki M, Tsujimoto K, Yamasaki H, Koyama AH. Antiviral activities of coffee extracts in vitro. Food Chem Toxicol. 2008;46:1919–24. https://doi.org/10.1016/j.fct.2008.01.031.
Clark KJ, Grant PG, Sarr AB, Belakere JR, Swaggerty CL, Phillips TD, Woode GN. An in vitro study of theaflavins extracted from black tea to neutralize bovine rotavirus and bovine coronavirus infections. Vet Microbiol. 1998;63:147–57. https://doi.org/10.1016/S0378-1135(98)00242-9.
••Agrawal A, Jain NK, Kumar N, Kulkarni GT. Molecular docking study to identify potential inhibitor of COVID-19 main protease enzyme: an In-silico approach. 2020. https://doi.org/10.26434/chemrxiv.12170904.v1This in silico study proposed that Oxyacanthine as a potential inhibitor of main protease enzyme of SARS-CoV-2 and could be explored for further drug development process.
Laskar MA, Choudhury MD. Search for therapeutics against COVID 19 targeting SARS-CoV-2 papain-like protease: an in silico study.2020; https://doi.org/10.21203/rs.3.rs-33294/v1
Murayama M, Tsujimoto K, Uozaki M, Katsuyama Y, Yamasaki H, Utsunomiya H, Koyama AH. Effect of caffeine on the multiplication of DNA and RNA viruses. Mol Med Report. 2008;1:251–5. https://doi.org/10.3892/mmr.1.2.251.
Sarma P, Shekhar N, Prajapat M, Avti P, Kaur H, Kumar S, Singh S, Kumar H, Prakash A, Dhibar DP, Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). J Biomol Struct Dynam. 2008;16:1–9. https://doi.org/10.1080/07391102.2020.1753580.
Weber JM, Ruzindana-Umunyana A, Imbeault L, Sircar S. Inhibition of adenovirus infection and adenain by green tea catechins. Antivi Res. 2003;58:167–73. https://doi.org/10.1016/S0166-3542(02)00212-7.
Bhowmik D, Nandi R, Jagadeesan R, Kumar N, Prakash A, Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Inf Gen Evol. 2020;84: 104451. https://doi.org/10.1016/j.meegid.2020.104451.
••Elzupir AO. Caffeine and caffeine-containing pharmaceuticals as promising inhibitors for 3-chymotrypsin-like protease of SARS-CoV-2. J Biomol Struct Dynam. 2020:1–8. https://doi.org/10.1080/07391102.2020.1835732. In this study, linagliptin and caffeine are recommended for COVID-19 treatment after in vitro, in vivo and clinical trial validation.
LeBlanc EV, Colpitts CC. The green tea catechin EGCG provides proof-of-concept for a pan-coronavirus entry inhibitor. bioRxiv. 2021. https://doi.org/10.1101/2021.06.21.449320.
Jang M, Park YI, Cha YE, Park R, Namkoong S, Lee JI, Park J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid Complem Altern Med. 2020;2020:17.
••Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020;20:1. https://doi.org/10.1080/07391102.2020.1766572. This study showed Oolonghomobisflavan as a potential bioactive molecule to act as an inhibitor for the Mpro of SARS-CoV-2.
Gottwalt B, Tadi P. Methylxanthines. StatPearls. 2021. https://www.ncbi.nlm.nih.gov/books/NBK559165/
Furge LL, Guengerich FP. Cytochrome P450 Enzymes in Drug Metabolism and Chemical Toxicology Biochemistry and Molecular Biology Education. 2006;34(2):66-74. https://doi.org/10.1002/bmb.2006.49403402066.
Acknowledgements
The authors acknowledge Shoolini University, Solan (India) for providing infrastructure support to conduct the research work. Authors also acknowledge the support provided by Yeast Biology Laboratory, School of Biotechnology, Shoolini University, Solan, India.
Author information
Authors and Affiliations
Contributions
Experiments were performed by RR, DS, BS and SS. OA, OAF, CNP helped in MD simulation. AS, VK and DJB helped in writing the manuscript and analysis of data. KD conceived the idea and provided guidance to execute the research project. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Human and Animal Rights and Informed Consent
The article does not contain any studies with human or animal subjects performed by any authors.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Natural Products: From Chemistry to Pharmacology
The editors would like to thank Dr. Heng Wee Tan for taking the time to review this manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rolta, R., Salaria, D., Sharma, B. et al. Methylxanthines as Potential Inhibitor of SARS-CoV-2: an In Silico Approach. Curr Pharmacol Rep 8, 149–170 (2022). https://doi.org/10.1007/s40495-021-00276-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40495-021-00276-3