Abstract
In simulation-based studies and analyses of epidemics, a major challenge lies in resolving the conflict between fidelity of models and the speed of their simulation. Another related challenge arises in dealing with the large number of what–if scenarios that need to be explored. Here, we describe new computational methods that together provide an approach to dealing with both challenges. A mesoscopic modeling approach is described that strikes a middle ground between macroscopic models based on coupled differential equations and microscopic models built on fine-grained behaviors at the individual entity level. The mesoscopic approach offers the ability to incorporate complex compositions of multiple layers of dynamics even while retaining the potential for aggregate behaviors at varying levels. It also is an excellent match to the accelerator-based architectures of modern computing platforms in which graphical processing units (GPUs) can be exploited for fast simulation via the parallel execution mode of single instruction multiple thread (SIMT). The challenge of simulating a large number of scenarios is addressed via a method of sharing model state and computation across a tree of what–if scenarios that are localized, incremental changes to a large base simulation. A combination of the mesoscopic modeling approach and the incremental what–if scenario tree evaluation has been implemented in the software on modern GPUs. Synthetic simulation scenarios are presented to demonstrate the computational characteristics of our approach. Results from the experiments with large population data, including USA, UK, and India, illustrate the modeling methodology and computational performance on thousands of synthetically generated what–if scenarios. Execution of our implementation scaled to 8192 GPUs of supercomputing platforms demonstrates the ability to rapidly evaluate what–if scenarios several orders of magnitude faster than the conventional methods.
Similar content being viewed by others
1 Introduction
The tremendous significance of analyzing and understanding the dynamics of epidemics is now abundantly clear in light of not only many major disease outbreaks of the past, but also with the most pronounced and consequential effects of COVID-19 worldwide2,25,26. Computer-based simulations are used for many purposes to deal with this problem, including prediction, confirmation, validation, exploration, enhancing our understanding, establishing limits, and so on. This brings greater emphasis to the need for creating the next generation of computational approaches to modeling and simulating epidemic dynamics12,13,19,23. This includes the need for advances in computer-based modeling capabilities by which the most important elements and behaviors are accurately composed and captured. It is now public knowledge that the epidemic-influenced world is one of the most complex systems humans have encountered so far, posing difficulties to the researchers in balancing the mutually opposing factors of model fidelity, simulation speed, and real-time evaluation. On a positive note, the challenge is also an excellent opportunity for the scientific community to explore, uncover, and offer newer concepts, technologies, and solutions to this class of problems and also consequently carry the advances to other domains as well for similar advancements.
1.1 Motivation
One of the advancements that the domain can use is in new modeling and simulation approaches that preserve the primary benefits of both the extremities, namely, macroscopic and microscopic modeling, while largely overcoming their shortcomings. The next advancement relates to the thorny problem faced by every modeling and simulation effort for complex systems: how can we effectively explore the vast parameter space of what–if analyses in which a large number of possibilities on the near-term time horizon are explored quickly as small, incremental variations of scenarios over the current, large state of the complex system6. The scenarios to be explored become numerous due to the multitude of factors at play, which include location-specific effects, behavioral effects, intervention measures, and so on2,9,28. On one hand, massive simulations of microscopic models cannot typically be run in large numbers of scenarios. On the other hand, macroscopic models are easily run, but the number of parameters is typically not as high as with higher resolution models1,3,8.
Apart from the number of scenarios, there is an additional challenge that is commonly faced, namely the problem of meeting real-time considerations. When simulation is used to explore the effects even as the situation is evolving, there is the additional pressure to evaluate as many scenarios as possible in a given amount of real time, so that well-informed decisions can be made quickly18,19,–20.
These considerations present new questions with respect to modeling and simulation technology4,13. Can we have a flexible middle ground between the two extremes of macroscopic and microscopic models? Can the scalability of macroscopic models be approached even while new models of dynamics are incorporated in arbitrary compositions? How can we modify a simulation scenario on the fly and create new what–if scenarios, even while the state trajectories in the original scenario continue to be evaluated over time, concurrently with the incremental what–if scenarios spawned from the original scenario? How can we let a scenario continue to run while sharing its state with the slightly changed what–if scenarios? Can we let the what–if scenarios reuse much of the original epidemic scenario, even as both continue to evolve in simulation time?
1.2 Contributions
In this paper, we focus on presenting new computational concepts and possibilities for next generation of epidemic simulations, by providing advancements on both aspects: fast base simulation and rapid exploration of what–if trees over the base simulation. As a middle ground, mesoscopic models represent an appropriate class in which the resolution and number of parameters are sufficiently high to capture a fair amount of phenomenological complexity, and thus warrant a good sweep of scenario parameters to improve the accuracy and assurance from simulation-based studies. Therefore, a mesoscopic simulation approach is described that offers the potential to serve as a continuum from macroscopic to microscopic levels, whose configurations can be customized based on data availability, desired modeling accuracy, targeted level of behavioral detail, and other such factors. Building over this mesoscopic framework, an incremental simulation approach based on what–if tree evolution is presented that offers new scaling capabilities that were not possible before in rapidly simulating thousands or millions of incrementally varied scenarios over a large domain of a base simulation2,25,26. The mesoscopic model can be used in the incremental what–if tree evaluation on state-of-the-art accelerated computing platforms including supercomputers that offer thousands of GPUs, and effectively exploit the single-instruction-multiple-data (SIMD) mode of high-performance parallel computing. The two approaches together form the basis for new computational ways of tackling the modeling and simulation problem in epidemic studies and decision systems.
Our focus here is on presenting novel computational approaches for fast simulation of large-scale population sizes and for rapid exploration of massive numbers of what–if scenarios. As a result, the epidemic dynamics do not specifically represent any particular configurations from real-life situations, although the general trends are validated to conform to the expect evolution from standard propagation models. The goal is to present the new computational methodologies that are now available from this research for domain scientists to explore and exploit towards real-life studies. Therefore, the approach and feasibility results presented here in scalable modeling and fast scenario exploration are envisioned to provide a leap in the simulation capabilities for analyzing epidemics and other complex systems.
A major challenge is the number of parameters, scenarios, and conditions at play. It is nearly impossible to run a simulation for every combination of parameters for scenarios30. Traditional simulations11 serve well with experiment design techniques to address the problem of sampling the state space. For significant advances, however, new paradigms need to be developed to advance the epidemiological simulation technology. Methods such as factorial experiment design and Latin square sampling are commonly used, along with recent methods for uncertainty quantification, but there is little support for incremental changes to large scenarios, and insufficient methodology to deal with large what–if trees in conjunction with mesoscopic simulations in an efficient manner. Compounding the problem, the additional dimension of real-time simulation and decision-making places a higher expectation of rapid evaluation of dynamically created scenarios. The what–if scenarios come in sequences and branches, forming a tree of evaluations. Also, the branches that need to be evaluated can be exogenous, based on new external inputs to the simulation based on evolving ground truth or influx of data. The updates may also appear in the form of internal simulation state-based conditions (e.g., a new event with a certain probability will arise only when the state satisfies certain conditions), which are difficult to schedule a priori in a conventional experiment design.
1.3 Organization
The rest of the paper is organized as follows. Our mesoscopic approach to the large, complex systems of epidemics is presented in Sect. 2. The computational framework to enable rapid exploration of massive trees of incremental what–if scenarios is described in Sect. 3. This is followed in Sect. 4 by a performance study using computational experiments on three basic country-scale models, with sequential (one node) performance as well as small-scale and large-scale parallel execution on up to 8192 GPUs. The paper is summarized and future work identified in Sect. 5.
2 Mesoscopic Approach to Large, Complex Systems
2.1 Conceptual Framework
The modeling spectrum for any large, complex system in general spans the space between two extremes with respect to the level of detail included per interacting entity in the system. In the epidemic case, the most numerous entities in the system are the individuals who are targets of the disease. Therefore, the two extremes for epidemic modeling span the two ends with respect to the level of detail captured in the model per individual. When the individuals are merely represented as counts of certain bins such as the total number of susceptible individuals and the number of infected individuals, such a model is a macroscopic model7. In a macroscopic model, there is little additional identity or distinguishing factor on an individual basis, but only aggregates are represented and tracked. In the other extreme, when each individual entity is demarcated and explicit state of its own that is tracked separately from that of other entities, such a model is a microscopic model10. Recently, mesoscale modeling has been used at local level or regional level for modeling and validating COVID-19 at the county level15.
Mesoscopic modeling can provide a good trade-off between fidelity and speed of simulation. At the right level of population density (number of individuals per grid cell), it can capture the dynamics of microscopic models with differential equations while providing the ease of composing many effects that can be varied at the level of each grid cell. At higher grid sizes, the population density can be reduced even further, potentially down to a few dozen people per grid cell. This can either be specified as a specialized, individual-level model or an aggregate, difference equation-based approximation to the high-resolution microscopic model.
2.2 Mapping the Model to SIMD Grid
For the highest resolution data partitioning of population to grid cells, it may be possible to change the mesoscopic model to a microscopic, individual-level model for more accurately capturing the dynamics. For example, in the case of UK, it is seen that the average population size per grid cell indeed approaches a single individual when the grid size is 8192 × 8192. The transformation of mesoscopic-to-microscopic model opens new research directions as future work.
Each cell in the grid represents an aggregate of population. Therefore, every grid cell executes the differential equations presented in the subsequent sections for the epidemic model, infection model, spatial model, and inter-entity interaction model. This makes the mesoscopic approach a strict superset of the macroscopic approach. Consequently, it is possible to take any macroscopic model and incorporate it into the mesoscopic model.
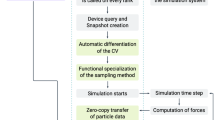
The power of this grid-based mesoscopic approach comes from its excellent alignment with the fast SIMD processing capabilities of modern GPUs. This match of the model to the GPUs offers three important benefits:
-
1.
This model can be executed on any desktop machine of a researcher or practitioner, making fast, large-scale simulations accessible to many important users without significant additional investments for computational support.
-
2.
The use of the GPU also makes it possible to offer interactive animations of the dynamics even as the simulations are executed. Since the GPU is also a rendering device, the latency between computation and visualization is minimized. Moreover, the visualization software layer is extremely light due to the native support provided by most GPU vendors, including standardized interfaces such as OpenGL.
-
3.
In the case of practitioners or researchers who have access to large parallel computing platforms, the GPU-based mesoscopic approach is perfectly suited to exploit the parallel system. Many large computational clusters, cloud computing platforms, or even supercomputers, offer GPU-based nodes. In fact, the top supercomputers of the world are currently ranked at the top exclusively because of their hardware design that is heavily GPU-based.
2.3 Epidemic Model
We illustrate our mesoscopic modeling approach using the parameters of an Ebola epidemic model, although the methodology is general and applicable to many other epidemics. For experimentation purposes, we use a few parameters and fitted values for an epidemic (Ebloa 2014) model22. The parameter values are shown in Table 1. We solve the system of linear equations using initial conditions with values corresponding to the countries. In particular, we assume that for the countries considered in this work, the initial exposed populations are both 10% of the aggregate ones.
Note that this configuration and settings are chosen only to exercise the computational framework, and hence are not meant as actual validated studies. Because any scenario-specific values can be varied by the user, the system is general purpose in nature and not limited to any specific model or components. Our focus here is limited to presenting the novel approach to modeling and simulation on current accelerated computing architectures.
2.4 Infection Model
We use a general metapopulation-based susceptible-exposed-infectious-recovered (SEIR) epidemic model5. We assume that the environment under consideration is divided into L location patches, which are geographic regions. Each patch is considered to be homogeneous and divided into four compartments where individuals are classified as:
-
S: Susceptible individuals, who can be infected;
-
E: Exposed individuals, who have been infected but not yet infectious;
-
I: Infectious cases in the community, who are capable of transmitting the disease;
-
R: Individuals removed from the chain of transmission (cured or dead and buried).
The number of people of each compartment in path i at some time t is denoted by \(S_i(t)\), \(E_i(t)\), \(I_i(t)\), and \(R_i(t)\), respectively for \(i=1,2,\ldots ,L\). The total number of people in patch i is denoted by \(N_i(t)=S_i(t)+E_i(t)+I_i(t)+R_i(t)\). The population will be constant during the outbreak.
The model takes into consideration the number of people infected due to direct contact with an infected individual and the number of people infected due to direct contact with latent individuals: \(\beta \frac{SI}{N}\). The individuals in the latent stage will eventually show the symptoms of the disease and enter into the infectious stage. This is denoted as \(\delta {E}\), where \(\delta\) is the per-capita infectious rate. In that case, \(\frac{1}{\delta }\) becomes the average time for a latent individual to become infectious. The recovery rate is denoted by \(\gamma I\), where \(\gamma\) is the per-capita recovery rate.
2.5 Spatial Mobility Model
We also model the behavior that individuals travel between the patches. The rates of travel of individuals between any two patches can be made to depend on the disease state. The disease states of individuals do not change during travel. The simplest travel pattern per time step is movement from any given grid cell to its immediate Moore neighborhood5,14,21. However, this can be customized with another mapping array for arbitrary connectivity, especially for modeling the non-local effects of air travel and long rail/highway-based interactions.
2.6 Consolidated Model
When the SEIR infection model is integrated with the spatial mobility model, each grid cell updates its counts based on the combined terms of compartments as described next.
Let \(m^{S}_{ij}\), \(m^{E}_{ij}\), \(m^{I}_{ij}\), and \(m^{R}_{ij}\) denote the travel rate from patch i to patch j of susceptible, exposed, infective, and recovered individuals, respectively, where \(m^{S}_{ii}=m^{E}_{ii}=m^{I}_{ii}=m^{R}_{ii}=0\). The travel rates among all the patches can be represented by matrices \(M^S=[m^{S}_{ij}]\), \(M^E=[m^{E}_{ij}]\), \(M^I=[m^{I}_{ij}]\), and \(M^R=[m^{R}_{ij}]\) where \(1 \le i,j \le L\)
This set of equations is mapped to each grid element. At every time step, the grid elements are updated concurrently in an SIMD fashion. Global statistics are computed periodically for visualization and output generation purposes.
2.7 Data Sources
The geographical population data per country is collected from the WorldPop dataset27 using the methods described in17. The population datasets are available in GeoTIFF format in 100 m and 1 km resolutions. We used the GDAL (Geospatial Data Abstraction Library) package to parse the dataset and extract the population counts per location patch. The data are used to generate intermediate population files per country for various grid sizes. We used grid sizes of 256, 512, 1024, 2048, 4096, and 8192 in the experiments. We used three different countries of varying area, density, and population counts: UK, USA, and India. A summary of the dataset is presented in Table 2.
Similar data sources can be used for pre-processing and reformatted to fit the mesoscale model. For large data sets, the pre-processing itself can be parallelized to reduce the amount of data preparation time. However, this needs to be done only once for each set of initial conditions. Typically, these input data do not change too frequently (typically, once or twice a year), which makes the pre-processing a small, fixed cost for the mesoscale approach.
2.8 Grid Size and Model Fidelity
Given a specific geographic domain and its population distribution, they can be mapped to a grid of desired size. Thus, for any given population size, the greater the size of the grid to which the population is mapped, the smaller is the number of people per grid cell, and therefore, the higher the resolution of representation. However, because the mesoscopic model is essentially a composition of multiple macroscopic compartments, the resolution must be such that macroscopic approach should remain valid within each grid cell. When the number of people per grid cell drops to a low value (unity or fraction), the macroscopic view of the grid cell no longer holds, and the mesoscopic model fails to provide correct results. Therefore, there is a trade-off between the amount of resolution and the validity of the model.
We have implemented this and empirically tested the phenomenon on varying grid sizes on representative large domain sets. Specifically, three countries, namely UK, USA, and India, were chosen to represent small, medium, and large population sizes, respectively. A summary of the dataset is presented in Table 3. We seek to answer the question: how do key epidemic observables vary as the size of the total grid is increased (that is, the resolution is increased from low to high)? Note that this is performed for the base simulation. The grid size is varied from 256 to 8192 by powers of two.
A verification of the mesoscopic SEIR infection measures reflects the expected trends from macroscopic differential equation-based curves. The results for different compartments as the grid size is varied are shown in Figs. 1, 2, and 3.
Because the values assigned to the infection parameters are fixed across all resolutions, the effective parameter values vary across the resolutions. The figures show that the model needs to be calibrated for any given grid size to match the desired behavior. Also, at a grid size of 4096 or higher, the population per cell for UK is too low to be valid for the mesoscopic model.
3 Evaluating Massive Trees of Incremental What–If Scenarios
In this section, we describe our approach to the problem of rapidly evaluating a large number of incremental simulations in a tree of what–if decisions relative to the base mesoscopic simulation.
3.1 Concepts and Terminology
The terminology underlying our what–if tree evaluation approach includes concepts of base simulation, what–if simulation, branches, and decision level, as described next.
-
Base simulation: This represents the full grid initialized and evolved as a complete, independent simulation, with no interference from the user.
-
What–if simulation: At any point during the evolution of the base simulation, a fraction (sub-rectangle) of the grid is overwritten with a new state that represents the new development to be explored. This represents an incremental change on top of the base simulation. To that extent, the what–if simulation shares the entire state of the base simulation except for this small portion that is changed to reflect the what–if configuration. Note that the what–if incremental simulation starts expanding its changed state beyond the initial fraction, because the grid cells affect their neighboring cells at each iteration of simulation time. There can be multiple ways the what–if scenarios can be initiated. New data feeds of ground truth can serve as sources of exogenous updates to the base simulation. Alternatively, new (probablistically generated) configurations can be introduced based on the current state of the simulation. These dynamically generated scenarios serve as endogenous updates that need to be evaluated for their effects in the simulated future.
-
Branches: At each decision point, there can be one or more variations that need to be explored in addition to the base simulation. This serves as a branch of what–if variants that are introduced into the what–if tree. For example, branches include choices for new outbreaks at one or more locations, new curfew or quarantining restrictions, new vaccination campaigns, etc. Note that these can be based either on real incidents and ground truth, or contemplated, planned actions and developments.
-
Decision level: The what–if scenarios can be spawned in a cumulative fashion as a sequence of what–if decisions to be made. Therefore, they result in a tree of scenarios, each being an incremental modification of its previous configuration, which ultimately is a union of changes of what–if scenarios from the leaves via their branches up to the base simulation. The level of the what–if is its distance from the original base simulation.
Note that an incremental what–if simulation could evolve over time to expand from a localized change to a globally different simulation from its base simulation. In other words, the incremental simulation for a what–if starts out with a small fraction of the global grid being set to the new what–if-defined state, but, starts expanding its evolution beyond the confines of that incremental state (because of neighborhood interactions). Eventually, the size of the incremental simulation thus expanding may reach the boundaries of the grid, at which point the incremental simulation can be considered a bonafide simulation of its own. This is because, from that point in simulation time, none of its state is shared with its ancestors in the what–if tree. In that situation, it need no more be maintained as an incremental simulation, but can be detached as its own original, full, base simulation in its own right, as though it was started with its own initial conditions.
3.2 Epidemic Scenarios
To study the impacts of various factors during the epidemic spread, we consider several well-studied scenarios. To evaluate the impact, a combination of these scenarios need to be tested16,24. Furthermore, the scenarios are also dependent on the type of geographical areas, such as cities, rural areas, mountains, rivers, etc. We present a brief overview of the scenarios considered in this paper (Figs. 4, 5, 6).
-
Outbreak Sometimes during an epidemic outbreak, some spatial regions become hot spot of epidemic outbreak. The geography and demography of those regions play an important role in understanding the control mechanism.
-
Spatial quarantine During an epidemic outbreak in a region, sometimes, travel restrictions have to be applied to control the spread of diseases. The effectiveness of a quarantine policy depends on the spatial movement properties, such as inter-cell transportation modes and associated delays. To more accurately model all transportation modes such as air travel, the size of the what–if sub-region may need to be expanded to include all cells in the region between the origins and destinations of travel. In this paper, we restrict the movement of individuals from and to a quarantined zone.
-
Hospitalization Many disease models include hospitalization as an intervention. We can introduce the variable Q to denote the infectious population being hospitalized, and the variable \(\alpha\) to denote the rate of hospitalization22. We assume that the hospitalized individuals share the same recovery probability with the normal infectious ones, but do not infect any exposed individual or susceptible one. With this approach, the base SEIR model is modified to include what–if scenarios for hospitalization as follows:
$$\begin{aligned} \begin{aligned} \frac{\mathrm{d}S_{i}}{\mathrm{d}t}&= -\frac{\beta }{\gamma }S_{i}(1-\alpha )I_{i} + \sum \limits _{j=1}^{L}\left( m^{S}_{ij}S_{j} - m^{S}_{ij}S_{i}\right) \\ \frac{\mathrm{d}E_{i}}{\mathrm{d}t}&= \frac{\beta }{\gamma }S_{i}(1-\alpha )I_{i} - \frac{\delta }{\gamma }{E_{i}} + \sum \limits _{j=1}^{L}\left( m^{E}_{ij}E_{j} - m^{E}_{ij}E_{i}\right) \\ \frac{\mathrm{d}I_{i}}{\mathrm{d}t}&= \frac{\delta }{\gamma }{E_{i}} -{I_{i}} + \sum \limits _{j=1}^{L}\left( m^{I}_{ij}I_{j} - m^{I}_{ij}I_{i}\right) \\ \frac{\mathrm{d}R_{i}}{\mathrm{d}t}&= {I_{i}} + \sum \limits _{j=1}^{L}\left( m^{R}_{ij}R_{j} - m^{R}_{ij}R_{i}\right) \\ \frac{\mathrm{d}Q_{i}}{\mathrm{d}t}&= \alpha I_{i} + \sum \limits _{j=1}^{L}\left( m^{Q}_{ij}Q_{j} - m^{Q}_{ij}Q_{i}\right) .\\ \end{aligned} \end{aligned}$$(2) -
Vaccination In the vaccination scenario, we apply vaccination to the susceptible individuals to become immune to the disease. To model vaccination, we can introduce another variable V(t) to be the number of individuals who have been vaccinated22. We let the vaccination rate be given as a function of time by \(\eta\). Thus, \(\eta\) is the number of individuals being vaccinated per unit time at time t. With this additional variant, the base SEIR model is modified to include what–if scenarios for vaccination as follows (this addition can be cumulative, together with the hospitalization what–if addition mentioned previously):
$$\begin{aligned} \begin{aligned} \frac{\mathrm{d}S_{i}}{\mathrm{d}t}&= -\beta \frac{S_{i}I_{i}}{N_{i}} - \eta + \sum \limits _{j=1}^{L}\left( m^{S}_{ij}S_{j} - m^{S}_{ij}S_{i}\right) \\ \frac{\mathrm{d}E_{i}}{\mathrm{d}t}&= \beta \frac{S_{i}I_{i}}{N_{i}} - \delta {E_{i}} + \sum \limits _{j=1}^{L}\left( m^{E}_{ij}E_{j} - m^{E}_{ij}E_{i}\right) \\ \frac{\mathrm{d}I_{i}}{\mathrm{d}t}&= \delta {E_{i}} -\delta {I_{i}} + \sum \limits _{j=1}^{L}\left( m^{I}_{ij}I_{j} - m^{I}_{ij}I_{i}\right) \\ \frac{\mathrm{d}R_{i}}{\mathrm{d}t}&= \delta {I_{i}} + \sum \limits _{j=1}^{L}\left( m^{R}_{ij}R_{j} - m^{R}_{ij}R_{i}\right) \\ \frac{\mathrm{d}V_{i}}{\mathrm{d}t}&= \eta + \sum \limits _{j=1}^{L}\left( m^{V}_{ij}V_{j} - m^{V}_{ij}V_{i}\right) .\\ \end{aligned} \end{aligned}$$(3) -
Logistics To tackle an epidemic disease, quick delivery of medical supplies and hospitalization are crucial. However, these might be delayed due to various forms of social and geographical factors. This aspect could potentially be separately modeled or included in the base SEIR model by changing the rate of movement of individuals in a region.
4 Computational Experiments
In the following computational experiments for a performance study, we use India geography and population data as the test case, with a grid size of \(2048\times 2048\). The experiments are run on a single computational node for a baseline sequential performance, and also on parallel computing platform with several GPUs. Two what–if scenario trees are used: a small scale what–if tree with 30,000 incremental simulation scenarios, and a large-scale what–if tree with tree size of approximately 350,000 scenarios.
4.1 Performance Experiment Configurations
4.1.1 Hardware and Software
We used a server with an NVIDIA Tesla V100 GPU with 16 GB RAM and Intel(R) Xeon(R) Silver 4110 CPU with 256 GB of host memory. The underlying operating system was UBUNTU 20.04. We used C++ for the software and CUDA 10.0 for the GPU.
For the large-scale parallel runs, we used a supercomputing system to conduct the multi-node experiments. The supercomputing system consists of 18,688 compute nodes, a total of 710 TB system memory, and Cray’s high-performance Gemini network. Each node hosts a 16-core AMD Opteron processor with 32 GB of host memory and an NVIDIA Tesla K20X GPU. Each GPU contains 2688 CUDA cores with 6 GB of device memory. The supercomputing system is based on CUDA 7.0 for the GPU, and a vendor-supplied native implementation of the Message Passing Interface (MPI) for inter-processor communication and synchronization.
4.1.2 Performance Parameters
The experimental runs are conducted for a number of simulation time steps to perform a sufficient mixture and reach of model dynamics. We used three key variables to spawn new what–if (incremental) simulation runs: (i) the fraction \(\Delta\) of the domain affected that defines each new what–if simulation; (ii) the number of what–if scenarios m per decision sequence, and the number of decisions k in sequence. We vary these parameters to evaluate the runtime performance of the system, using a value of \(\Delta =10^{-3}\) on each spatial dimension.
The spatial grid dimension \(W \times H\) with width W and height H of all our simulation experiments is set to \(2048 \times 2048\). Larger dimensions result in higher resolutions and larger domains, for which what–if simulation would perform even better. For this grid size and \(\Delta =10^{-3}\), each what–if scenario’s initial dimensions as \(\sqrt{2048\times 2048\times 10^{-3}}\approx 64\). Hence, each what–if simulation starts with an incrementally changed domain size of \(64 \times 64\), which will be modified with scenario-specific spatial data (e.g., new infections centered at the chosen location). In the experiments regarding performance evaluation, we used the country data for India.
The simulation model tracks the propagation dynamics based on the SEIR model specified in Sect. 2. The incremental simulations are spawned based on a variety of what–if scenarios, such as new outbreaks (grids with increased infected count), quarantines (restricted spatial movement), vaccination (reducing susceptible counts), and hospitalization (increasing recovered counts).
4.1.3 Performance Metrics
As the measure of computational effectiveness of our what–if tree evaluation framework, we use a notion of speed-up that is different from the concept of parallel speed-up traditionally used in parallel computing. In parallel computing, the (strong scaling) speed-up is the factor of reduction in total computational time when multiple processors are used, relative to the time taken when only one processor is used. Here, we use a different notion, because the parallel speed-up is not applicable.
Whether one processor is used or multiple processors are used, the total time taken by the traditional simulation techniques to complete M what–if scenarios is \(M \times T_{\mathrm{C}}\), where \(T_{\mathrm{C}}\) is the time taken to complete one simulation. Note that, in normal simulation approaches, each what–if scenario essentially becomes a full simulation of its own. In our approach, only the base simulation is by default a full simulation, but the what–if scenarios are only incremental in nature, so consume time to simulate only a fraction \(\Delta\) of the original domain. Therefore, the time taken by our framework is significantly less than that of normal (replicated) simulations for each what–if scenario. A more detailed analysis of this complexity is available in our previous work29. Based on this notion, the speed-up is defined as the factor of reduction in time using our what–if incremental scenario approach compared to fully replicated runs. Note that the savings from experiment design methods apply equally to both traditional replication approach as well as to our approach, because any scenarios that can be avoided using some such design can be equally applied to our framework as well. As such, this is a robust measure with respect to comparing scenario evaluation via full simulation versus our what–if tree evaluation method.
4.2 Single-Node Performance Results
In this set of experiments, all simulations are performed on a single computational node. In this single-node execution, all the incremental what–if simulations are performed in conjunction with the base simulation on a single computing node with only a single GPU. To evaluate the performance, we run multiple scenarios with varying number \(N_{B/L}\) of what–if branches per decision level and number of decision levels \(N_{L}\). Table 4 gives the aggregate number of what–if simulations at each level, during what–if executions. The simulation was run for 100 timesteps and \(N_{L}=5\) and \(N_{B/L}\) was varied from 1 to 6. We used a value of \(\Delta =10^{-3}\) for this experiment. Table 5 shows the speed-up from incremental simulation. We can see orders of magnitude in speed-up gain, peaking at 350 when the number of what–if scenarios handled is 1555.
4.3 Parallel Execution Performance Results
We evaluated the scaling behavior using two experiments: (a) small scale and (b) medium scale. In the small-scale experiments, the simulation was run for 100 time steps, with \(N_{B/L}=3\) and \(N_{L}=10\), resulting in the spawning of 29, 524 what–if simulations and, the same scenario was run on 32, 48, 64, 96, 128, 256, 512, and 1024 GPUs. In the large-scale experiment, the simulation was run for 100 time steps, with \(N_{B/L}=4\) and \(N_{L}=10\), resulting in the spawning of 349, 225 what–if simulations and it was executed on 512, 1024, 2048, 4096, and 8192 GPUs.
The summaries of the results for the small- and large-scale simulations are tabulated in Tables 6 and 7, respectively. From the experimental results, we can observe that the performance gain increases significantly with an increase of the number of nodes. Using up to 8192 nodes (GPUs), we can achieve a performance gain of approximately 75 K for approximately 350 K what–if simulations.
5 Summary and Future Work
A mesoscale modeling approach is presented that appears suitable for modeling epidemics at large scales for first-order metrics, and well suited for exploiting the GPU platforms. We have also presented a framework for generating and evaluating massive what–if scenarios, scalable from single machine to large supercomputing platforms.
It is now conceivable to rapidly evaluate millions of what–if scenarios to adequately cover the parameter space and aid informed decision-making in real time in evolving epidemics. The mesoscopic modeling approach presented here provides an effective way to exploit the computational power of GPU hardware technology. In some of the largest experiments, the parallel computational runs show the feasibility to utilize thousands of GPUs to explore what–if trees containing many hundreds of thousands of decision sequences in a matter of minutes. The results presented here represent some of the largest and fastest what–if simulations reported in the literature. In the largest case, nearly 350,000 what–if scenarios were executed on 8192 GPUs in about 2 min of wall-clock time. The same system is also usable on commodity desktop computers for local and regional-scale simulation and analyses.
To use this model for a given geographical population density and grid size, the parameters need to be calibrated to maintain the right balance of macroscopic versus microscopic level of models. As shown in Sect. 2, the exact global counts of SEIR compartments are dependent on the grid size and constants used in the model. This needs to be studied to provide the mathematical methodology needed to partition a macroscopic model into multiple sub-macroscopic models, so that they can be mapped to the grid. Another class of shortcomings of our approach lie in the mapping from the geographical domain to the grid. Since the representation is a direct mapping, domains with contiguous land will perform better, whereas gaps in inhabitation on the land or presence of water bodies or other geographical separations will waste the grid cells in the GPU memory and computation. Another shortcoming is the availability of accurate data to initialize the simulation state in terms of the mesoscopic grid. Overall, there are multiple factors that need to be addressed before our approach can be readily used by actual decision-makers or practitioners. For instance, verification, validation, and accreditation processes will need to be undertaken to customize it for real-life use. Accordingly, our focus in this paper has been to first present the computational advancement that the mesoscopic representation provides and showing the potential for evaluating massive trees of what–if scenarios as incremental simulations executed on state-of-the-art GPU-based computing platforms.
References
Adiga A, Dubhashi D, Lewis B, Marathe M, Venkatramanan S, Vullikanti A (2020) Mathematical models for Covid-19 pandemic: a comparative analysis. J Indian Inst Sci 100:793–807. https://doi.org/10.1007/s41745-020-00200-6
Adiga A, Wang L, Sadilek A, Tendulkar A, Venkatramanan S, Vullikanti A, Aggarwal G, Talekar A, Ben X, Chen J, Lewis B, Swarup S, Tambe M, Marathe M (2020) Interplay of global multi-scale human mobility, social distancing, government interventions, and covid-19 dynamics. medRxiv. https://doi.org/10.1101/2020.06.05.20123760
Adiga A, Wang L, Hurt B, Peddireddy A, Porebski P, Venkatramanan S, Lewis B, Marathe M (2021) All models are useful: Bayesian ensembling for robust high resolution covid-19 forecasting. medRxiv. https://doi.org/10.1101/2021.03.12.21253495
Ajelli M, Gonçalves B, Balcan D, Colizza V, Hu H, Ramasco JJ, Merler S, Vespignani A (2010) Comparing large-scale computational approaches to epidemic modeling: agent-based versus structured metapopulation models. BMC Infect Dis 10(1):1–13
Arino J, Van den Driessche P (2006) Metapopulation epidemic models. a survey. Fields Inst Commun 48:1–13
Bradley E, Marathe M, Moses M, Gropp WD, Lopresti D (2020) Pandemic informatics: preparation, robustness, and resilience; Vaccine distribution, logistics, and prioritization; and Variants of concern. arXiv:2012.09300
Brauer F (2008) Compartmental models in epidemiology. In: Mathematical epidemiology. Springer, pp 19–79
Calvetti D, Hoover AP, Rose J, Somersalo E (2020) Metapopulation network models for understanding, predicting, and managing the coronavirus disease covid-19. Front Phys 8:261
Chang S, Wilson ML, Lewis B, Mehrab Z, Dudakiya KK, Pierson E, Koh PW, Gerardin J, Redbird B, Grusky D, Marathe M, Leskovec J (2021) Supporting covid-19 policy response with large-scale mobility-based modeling. medRxiv. https://doi.org/10.1101/2021.03.20.21254022
Eubank S, Guclu H, Kumar VA, Marathe MV, Srinivasan A, Toroczkai Z, Wang N (2004) Modelling disease outbreaks in realistic urban social networks. Nature 429(6988):180–184
Fujimoto RM (2000) Parallel and distributed simulation systems. Wiley-Interscience, New York
Gauvin L, Panisson A, Barrat A, Cattuto C (2015) Revealing latent factors of temporal networks for mesoscale intervention in epidemic spread. arXiv:150102758
Hemmert KS, Bair R, Bhatele A, Groves T, Hammond SD, Levenhagen MJ, Mubarak M, Pakin S, Ross R, Wilke JJ, Georgakoudis G (2019) System-level architecture simulation for exascale: challenges and opportunities. https://www.osti.gov/biblio/1639211
Kelly MR Jr, Tien JH, Eisenberg MC, Lenhart S (2016) The impact of spatial arrangements on epidemic disease dynamics and intervention strategies. J Biol Dyn 10(1):222–249
Kergaßner A, Burkhardt C, Lippold D, Kergaßner M, Pflug L, Budday D, Steinmann P, Budday S (2020) Memory-based meso-scale modeling of covid-19: county-resolved timelines in Germany. Comput Mech. https://doi.org/10.1007/s00466-020-01883-5
Liu S, Poccia S, Candan KS, Chowell G, Sapino ML (2016) epiDMS: data management and analytics for decision-making from epidemic spread simulation ensembles. J Infect Dis 214(4):S427–S432. https://doi.org/10.1093/infdis/jiw305
Lloyd CT, Chamberlain H, Kerr D, Yetman G, Pistolesi L, Stevens FR, Gaughan AE, Nieves JJ, Hornby G, MacManus K, Sinha P, Bondarenko M, Sorichetta A, Tatem AJ (2019) Global spatio-temporally harmonised datasets for producing high-resolution gridded population distribution datasets. Big Earth Data 3(2):108–139. https://doi.org/10.1080/20964471.2019.1625151
López L, Rodo X (2021) A modified Seir model to predict the covid-19 outbreak in Spain and Italy: simulating control scenarios and multi-scale epidemics. Results Phys 21:103746
Minutoli M, Sambaturu P, Halappanavar M, Tumeo A, Kalyanaraman A, Vullikanti A (2020) Preempt: scalable epidemic interventions using submodular optimization on multi-gpu systems. In: 2020 SC20: international conference for high performance computing, storage and analysis (SC). IEEE Computer Society, Networking, pp 765–779
Mishra S, Steen R, Gerbase A, Lo YR, Boily MC (2012) Impact of high-risk sex and focused interventions in heterosexual hiv epidemics: a systematic review of mathematical models. PLoS One 7(11):e50691
Ni S, Weng W (2009) Impact of travel patterns on epidemic dynamics in heterogeneous spatial metapopulation networks. Phys Rev E 79(1):016111
Ouyang X, Son S, Yu K (2015) Modeling the spread of ebola. Mathematical contest in modeling. https://sites.math.washington.edu/~morrow/mcm/mcm15/38725paper.pdf
Perumalla KS, Seal SK (2012) Discrete event modeling and massively parallel execution of epidemic outbreak phenomena. Simulation 88(7):768–783
Rivers CM, Lofgren ET, Marathe M, Eubank S, Lewis BL (2014) Modeling the impact of interventions on an epidemic of Ebola in Sierra Leone and Liberia. PLoS Curr. https://doi.org/10.1371/currents.outbreaks.4d41fe5d6c05e9df30ddce33c66d084c
Singer G, Marudi M (2020) Ordinal decision-tree-based ensemble approaches: the case of controlling the daily local growth rate of the covid-19 epidemic. Entropy. https://doi.org/10.3390/e22080871
St-Onge G, Thibeault V, Allard A, Dubé LJ, Hébert-Dufresne L (2021) Social confinement and mesoscopic localization of epidemics on networks. Phys Rev Lett 126(9):098301
WorldPop (2021) Population counts. https://www.worldpop.org/geodata/listing?id=29
Wu N, Ben X, Green B, Rough K, Venkatramanan S, Marathe M, Eastham P, Sadilek A, O’Banion S (2020) Predicting onset of covid-19 with mobility-augmented seir model. medRxiv. https://doi.org/10.1101/2020.07.27.20159996
Yoginath SB, Perumalla KS (2018) Scalable cloning on large-scale gpu platforms with application to time-stepped simulations on grids. ACM Trans Model Comput Simul 28(1):5:1–5:26. https://doi.org/10.1145/3158669
Zhang T, Lees M, Kwoh CK, Fu X, Lee GKK, Goh RSM (2012) A contact-network-based simulation model for evaluating interventions under what-if scenarios in epidemic. In: Proceedings of the 2012 winter simulation conference (WSC). IEEE, pp 1–12
Acknowledgements
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Funding
Funding was provided by Oak Ridge National Laboratory (Internal Research and Development).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Perumalla, K.S., Alam, M. Mesoscopic Modeling and Rapid Simulation of Incremental Changes in Epidemic Scenarios on GPUs. J Indian Inst Sci 101, 357–370 (2021). https://doi.org/10.1007/s41745-021-00253-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41745-021-00253-1