Abstract
Astroviruses are considered the cause of gastroenteritis in humans and animals. Studies in recent years show avian astroviruses are also associated with duckling hepatitis, gosling gout, and chicken nephritis. In this study, a GAstV strain, designated as JS2019/China, was detected in dead goslings from a commercial goose farm in Jiangsu province of China. Viral strain was proliferated in goose embryos and sequence analysis showed the isolated strain had a classical structure arrangement and a series of conserved regions compared with other GAstVs. Sequence comparison and phylogenetic analysis of whole genome and ORF2 revealed that JS2019/China belongs to the GAstV-1 group, which consists of most of the GAstV strains. Amino acid analysis indicated that some mutants might have an impact on viral protease capacity, such as V505I and K736E of ORF1a and T107I, F342S, and S606P of ORF2. Taken together, a novel GAstV strain was isolated and genomic analysis and protein polymorphism analysis indicated that some amino acid mutants might affect the viral virulence.
Similar content being viewed by others
Introduction
Astroviruses (AstV) are single-stranded, positive-sense, non-enveloped RNA viruses, which cause infection in a wide range of mammals and birds via the fecal–oral route [1, 2]. With the development of next-generation sequencing and vial surveillance, many more AstV-infected animal hosts have been discovered in recent years [2]. Avian astroviruses, AAstVs, comprise duck astrovirus 1, turkey astrovirus 1 and 2, and avian nephritis virus of chickens [3]. Although great efforts have been made to classified AAstVs according to the host of origin and the amino acid sequences of viral capsid protein (https://talk.ictvonline.org/), a lot of newly reported viruses have not yet been classified officially, such as duck astrovirus CPH [4] and YP2 and astroviruses of chicken origin [5] and goose origin [6, 7]. Emerging infectious disease of goslings broke out in many provinces of China since 2017, such as Shandong, Anhui, Guangdong, and Henan [7,8,9,10,11,12]. The clinical symptoms of infected goslings include gout and visceral hemorrhage and the pathogen of this disease was finally confirmed to be a new type of goose astrovirus (GAstV) [13].
GAstVs belong to the Astroviridae family and their RNAs are almost 7.0 kb in length, which consists of 5′-untranslated region (UTR), ORF1a, ORF1b, ORF2, 3′UTR, and a poly (A) tail [14, 15]. ORF1a and ORF1b encode non-structural proteins, which are responsible for the transcription and replication of the virus, and ORF2 encodes capsid protein [1, 16, 17]. ORF1b appeared to be the most conserved region, indicating that RNA-dependent RNA polymerase (RdRp), encoded by ORF1b, might be the most vital element for astrovirus replication [10].
In this study, a new goose astrovirus was isolated from dead goslings with gout and flu symptoms. The genomic characteristics and genetic relationships were analyzed between the newly isolated strain and other reference strains.
Materials and methods
Sample collection and viral nucleic acids detection
Samples of 5 dead goslings with gout and flu clinical signs were collected from a commercial goose farm in Jiangsu province of China in 2019. The liver, kidney, and small intestine tissue from different dead goslings were pooled and total RNA was extracted. cDNA was reversely synthesized using Hifair® II 1st Strand cDNA Synthesis Kit (Yeasen) according to the manufacturer’s instructions and tested by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) assay for the presence of potential viral RNA. qRT–PCR was performed to test particular viral genes using pairs of specific gene primers for GPV, TMUV, GRV, and GAstV (Table 1).
Virus isolation and identification
Liver, kidney, and small intestine samples from different dead goslings were collected and homogenized in sterile phosphate-buffered saline (PBS, pH 7.2) to give a 20% suspension (w/v), and centrifuged at 8000 × g at 4 °C for 10 min. Supernatants were filtered through a 0.22-μm syringe-driven filter. The filtrate was inoculated into the allantoic cavity of 10-day-old goose embryos (0.1 ml/egg). The allantoic fluid and embryoid body were harvested aseptically at 7dpi.
Viral genome sequencing
To determine the full-length nucleotide sequences of the virus, RACE-PCR was performed using RNA extracted from the allantoic fluid and embryoid body with a SMARTer®RACE 5′/3′Kit (Takaka Bio China, Inc.). The complete genome sequence was divided into five fragments based on the conserved regions according to the multiple sequence alignment using the MegAlign software. The primer sequences for different fragments are listed in Table 2. PCR products were cloned into the pMD19-T vector (Takara) and sequenced. Genome recombinant analysis was performed by DNAMAN 5.0 software.
Genomic characterization and phylogenetic analysis
Genome ORFs prediction was conducted by ORF finder in NCBI ( https://www.ncbi.nlm.nih.gov/orffinder/). The prediction of domains, repeats, motifs, and features of three ORFs was conducted by SMART tool (Simple Modular Architecture Research Tool) (http://smart.embl.de/). The nuclear localization signal prediction was conducted by NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/). The prediction of the potential viral protein genome-linked (Vpg) site was conducted by FoldIndex (https://fold.weizmann.ac.il/fldbin/findex). Conserved domains of the viral sequence analyses were conducted by CD search in NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Nucleic acid and amino acid sequences’ identity analysis were conducted by Nucleotide and Protein BLAST in GenBank. The multiple amino acid sequences of three ORFs of GAstVs were aligned by the Clustal W method to analyze the mutations in JS2019/China.
A phylogenetic tree of the complete genome of GAstV JS2019/China and other reference strains was produced using the maximum likelihood method and phylogenetic trees of three individual viral proteins were produced using the neighbor-joining method and bootstrap test of 1000 replicates in MEGA 5.0 software (MEGA, Pennsylvania State University, University Park, PA).
Protein structure predictions of capsid spike were performed. Secondary structure (alpha-helices and beta-sheets) were predicted by PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). Tertiary structure was predicted by phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2). Three-dimensional structures were analyzed and displayed using PyMOL software.
Results
Virus detection and isolation
The tissue samples were determined by RT–PCR/qRT-PCR for several common viruses, such as goose parvovirus, duck Tembusu virus, goose reovirus, and goose astrovirus. It is noteworthy that all of them showed negative results except for goose astrovirus (GAstV) (Fig. 1a).
The pathogenicity and detection of GAstV JS2019/China to goose embryos. a Detection the isolated virus by RT PCR with specific primers for GPV, TMUV, GRV, and GAstV. b Presence of varying degrees of hemorrhage on the goose embryo bodies infected with newly identified GAstV at 7dpi. c Detection of the GAstV in allantoic fluid from infected goose embryos. Fragments were amplified with the template cDNA, which were reverse transcripted from viral RNA. F1, the first generation of GAstV; F2, the second generation of GAstV. d Different DNA fragments of GAstV amplified from isolated virus
For isolating GAstV, healthy goose embryos were inoculated with the virus filtrate and after two passages, all the embryos were survived whereas different levels of hemorrhage of embryo bodies were observed at 7 dpi (Fig. 1b). On the contrary, a similar method was applied to healthy duck embryos and the virus was not propagated successfully, suggesting that the virus strain was specific to the goose.
Complete genome characteristics analysis
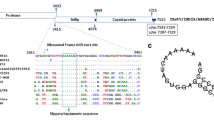
The virus strain isolate in this study was designated as JS2019/China. The full genome of JS2019/China is 7176 nucleic acids in length with 3 overlapping ORFs (ORF1a, 3105 nucleic acids; ORF1b, 1551 nucleic acids; ORF2, 2115 nucleic acids), and had a 5′-untranslated region (5′UTR) of 169 nucleic acids and a 3′UTR of 203 nucleic acids and 22 nucleic acids poly(A) tail (Fig. 2a). Sequences’ data have been submitted to Genbank and the accession number is MZ540211.
Predicted genome structure of isolated GAstV JS2019/China. a Genomic organization and functional domains of three ORFs. The complete genome showed a classical structure arrangement as 5′UTR (169 nt)-ORF1a (3105 nt)-ORF1b (1551 nt)-ORF2 (2115 nt)-3′UTR (203 nt)-poly(A)(22 nt). The functional domains of three ORFs were indicated by colored square. CC, coiled-coil domain; TM, transmembrane helical domain; PRO, protease motif; NLS, nuclear localization signal; Vpg, genome-linked viral protein; Znf, zinc finger-like motif; RdRP, RNA-dependent RNA polymerase. b The heptamer ribosomal frameshift signal (AAAAAAC) as a ribosome translocation signal in front of a “H-type pseudoknot” motif structure. c A conserved stem-loop-ll-like motif (s2m) located at 3′UTR of AAstVs
ORF1a (170–3274 bp) encoded a non-structural polyprotein of 1034 amino acid(aa), including a trypsin-like peptidase domain (504aa-656aa), a nuclear localization signal (723KKKGKTKKTAR733), two coiled-coil domains (84–127aa, 685–712aa), four potential transmembrane domains (181–198aa, 334–356aa, 371–393aa, 405–427aa), and a conserved zinc finger-like DNA-binding motif. The VPg site was predicted at the third disordered region from 699 to 761 aa, which contained the potential NLS coding site.
ORF1b (3265–4815 bp) encoded the RNA-dependent RNA polymerase (RdRP) of 383 aa, which had highly conserved motifs like G327NPSGQYSTTVDNN, Y377GDD, and F405GMWVK. A heptamer ribosomal frameshift signal (AAAAAAC) (3265–3271), located in the linker region between ORF1a and ORF1b, was regarded as a ribosome translocation signal (RFS) followed by a stem-loop structure (Fig. 2b). ORF2 (4834–6948 bp), the most variable region, encoded the precursor of capsid protein.
A stem-loop-ll-like motif (s2m) (Fig. 2c) located in the 3′UTR region may play a vital role in viral replication and natural recombination; it was reported to be conserved in astroviruses, coronaviruses, avian infectious bronchitis virus, and other non-related viruses [20].
Genetic and phylogenetic analysis
To investigate the genetic relationships, we compared the nucleotide sequences of the whole genome and ORFs between the newly isolated strain and other species of avian astroviruses. As seen in Table 3, the shared identities of ORFs were 26.8–60.0% (ORF1a), 51.9–68.0% (ORF1b), and 10.1–55.9% (ORF2). It is noteworthy that the RdRp domain of ORF1b is highly homologous to other species of AAstVs and the N-terminal capsid of ORF2 is highly homologous to TAstV-2 and DAstV-1.
Genetic analysis of ORF2 amino acid sequence showed that the mean amino acid distances (p-distances) were 0.410–0.719 between JS2019/China and members of the classified Avastrovirus species. Based on the species demarcation criteria (between species: 0.576–0.742, within species: 0.204–0.284), we demonstrated JS2019/China could be classified in the genus Avastrovirus. The genetic distance of ORF2 between JS2019/China, TAstV-2/3, and DAstV-1 was 0.403–0.410, indicating that they were highly related but different from each other. Meanwhile, the genetic distance between JS2019/China and other GAstV strains indicated that GAstVs could be divided into two groups. As shown in Table 3, the p-distance among JS2019/China and GAstV strain (MN 127,956) was just 0.004, demonstrating the identified astrovirus belongs to GAstV-1 group.
To further investigate the evolutionary history and relationship between newly isolated GAstV strain and other 49 reference AAstVs available in the GenBank database, the phylogenetic tree was constructed based on complete genome sequences and individual proteins (ORF1a, ORF1b, and ORF2) with MEGA 5.0 software. As shown in Fig. 3a, the phylogenetic tree of complete genomes indicated that GAstVs could be separated into GAstV-1 group and GAstV-2 group, proposing they were derived from different evolutionary histories. The JS2019/China was clustered into GAstV-1 group, which had a high identity (99.6%) to the GAstV strain of Guangdong (MN 127,956) as mentioned above and close to TAstV-2 and DAstV-3. Based on the phylogenetic trees of individual proteins using the N-J method, three of the ORFs were all assigned to the GAstV-1 group (Fig. 3b-d). ORF1b had a high identity (99.2–100%) to other GAstVs, while the identities of ORF1a and ORF2 were 97.8–99.7% and 97.7–99.6%, respectively. These results indicated that ORF1a and ORF2 were variable regions compared with other GAstV strains, and ORF1b was the most conserved region encoding a vital element for astrovirus replication as reported by Zhang [10]. Particularly, ORF1a and ORF1b were closely associated with TAstV-2 and DAstV-3, while ORF2 was closely associated with TAstV-2 and DAstV-1. These findings further indicated that different species of AAstVs had complex cross-species transmission and recombination. Based on the analysis results mentioned above, JS2019/China was classified into GAstV group-1 of Avastrovirus-3.
Phylogenetic analysis of complete genome and three ORFs. a Phylogenetic tree based on the complete genome of GAstV JS2019/China strain with other 49 reference strains from diverse avian species was constructed in MEGA 5.0 using the Maximum-Likelihood method with 1,000 bootstrap replicates. The isolated GAstV strain was indicated by black shaded triangle “▲”. b–d Phylogenetic trees were constructed in MEGA 5.0 using the neighbor-joining method, based on the amino acid sequence of ORF1a, ORF1b, and ORF2 GAstV JS2019/China strain with other reference strains same as above AAstVs, respectively
Amino acid polymorphism analysis
To further observe the divergences in ORFs among the newly isolated strain and other GAstV strains, we aligned the amino acid sequences of three ORFs and found the different amino acids (Table 4). The results demonstrated that ORF1b appeared to be a conserved domain with rare mutations. While ORF1a and ORF2 had more polymorphisms compared with other GAstV strains. The L149M and K736E mutations were located in ORF1a and the trypsin-like peptidase domain had a variable site at 505. Notably, the T107I, F342S, and S606P mutations of ORF2 are located in capsid N domain and capsid p2 domain, respectively. Meanwhile, these mutations of ORF1a and ORF2 changed the amino acid polarity. The impact of these mutations on viral protease capacity and antigenicity needs further investigation.
Capsid spike domain’s comparison of JS2019/China and HAstV-8
As the crystal structure of the GAstV has not been reported, we employed the Phyre2 website to predict the capsid spike structure of JS2019/China and analyzed the results with published crystal structures of the TAstV-2 and HAstV-8 spike domain [21, 22] (Fig. 4a-b). The capsid spike structure of JS2019/China is composed of two domains, namely the S domain (residues 66 to 249) and the P1 domain (residues 250 to 396). The S domain was mostly conserved and usually including a typical jelly-roll β-barrel fold (strands 1 to 8) [16, 23, 24]. The P1 domain contains partly conserved motifs, such as the α6 loop, the β12-α8 loop, and the β13 loop. As shown in the Fig. 4c, although the amino acid sequence of HAstV-8 (GenBank accession number AF260508) has a low identity with JS2019/China, the capsid spike structure of GAstV JS2019/China is much more similar with HAstV-8 compared to TAstV-2. JS2019/China has two α-helices different from HAstV-8, indicating that the capsid protein is specifically. The C terminal of the P1 domain was a similar structure, which was located externally near the S-P1 domain interface and was expected to connect the P2 domain [25].
Structure of GAstV JS2019/China. a Secondary structure assignment of capsid spike of JS2019/China, 66aa-396aa. α, α-helices; β, β-strands. b Predicted capsid spike structure of GAstV JS2019/China. c The alignment between predicted structure and HAstV-8 (PDB entries 5ibv). The HAstV-8 structure is colored in blue. GAstV JS2019/China structures were helix in red, beta sheet in yellow, and loop in aquamarine
Discussion
The goose industry is an important component of the poultry industry in China and avian gout is a major metabolic disease caused by avian astrovirus. In our study, we detected and isolated a novel GAstV strain and it was propagated successfully in goose embryos, but not duck embryos. The complete genome showed a classical structure arrangement. Genetic and phylogenetic analysis manifested that the JS2019/China strain was classified into the GAstV-1 group, which was closely related to TAstV-2 and DAstV-1. The sequence analysis also showed a high identity between the isolated strain and GAstV Guangdong strain. Perhaps, the development of modern transportation brings more challenges for the prevention and control of virus infection. As no antiserum or detection kit is specifically designed for GAstV currently [14], so our analysis of the mutations located in ORF2 may provide clues for understanding the pathogenic mechanism and helping for the effective vaccine development. Furthermore, the P1 domain, especially the highly conserved motifs mentioned above, might be immunogenic and contain neutralization epitopes as reported about HAstV-8. These predictions will be emphatically explored in the future researches.
References
Cortez V et al (2017) Astrovirus biology and pathogenesis. Annu Rev Virol 4(1):327–348
Karlsson EA et al (2015) Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog 11(11):e1005225
Koci MD, Schultz-Cherry S (2002) Avian astroviruses. Avian Pathol 31(3):213–227
Liu N, Wang F, Zhang D (2014) Complete sequence of a novel duck astrovirus. Arch Virol 159(10):2823–2827
Sajewicz-Krukowska J, Domanska-Blicharz K (2016) Nearly full-length genome sequence of a novel astrovirus isolated from chickens with “white chicks” condition. Arch Virol 161(9):2581–2587
Yuan X et al (2019) Genome analysis of newly emerging goose-origin nephrotic astrovirus in China reveals it belongs to a novel genetically distinct astrovirus. Infect Genet Evol 67:1–6
Zhang Q et al (2018) Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg Microbes Infect 7(1):71
Wei F et al (2020) Isolation and characterization of a duck-origin goose astrovirus in China. Emerg Microbes Infect 9(1):1046–1054
Wang Y et al (2020) Genomic and phylogenetic characteristics of a novel goose astrovirus in Anhui Province Central-Eastern China. Gene 756:144898
Zhang X, Deng T, Song Y, Liu J, Jiang Z, Peng Z, Guo Y, Yang L, Qiao H, Xia Y, Li X, Wang Z, Bian C (2022) Identification and genomic characterization of emerging goose astrovirus in central China, 2020. Transbound Emerg Dis 69(3):1046–1055. https://doi.org/10.1111/tbed.14060
Li JY et al (2021) Isolation and evolutionary analyses of gout-associated goose astrovirus causing disease in experimentally infected chickens. Poult Sci 100(2):543–552
Wang AP et al (2021) Isolation and characterization of a goose astrovirus 1 strain causing fatal gout in goslings, China. Poult Sci 100(11):101432
Niu X et al (2018) Novel goose astrovirus associated gout in gosling. China Vet Microbiol 220:53–56
Pantin-Jackwood MJ et al (2011) Molecular characterization of avian astroviruses. Arch Virol 156(2):235–244
Xue J et al (2020) Complete genome sequence and phylogenetic analysis of novel avastroviruses circulating in China from 2016 to 2018. Virus Res 278:197858
Arias CF, DuBois RM (2017) The astrovirus capsid: a review. Viruses 9(1):15. https://doi.org/10.3390/v9010015
Lewis TL et al (1994) Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J Virol 68(1):77–83
Corrand L et al (2011) Pathological and epidemiological significance of goose haemorrhagic polyomavirus infection in ducks. Avian Pathol 40(4):355–360
Zhang X et al (2018) An emerging novel goose astrovirus associated with gosling gout disease, China. Emerg Microbes Infect 7(1):152
Jonassen CM, Jonassen TO, Grinde B (1998) A common RNA motif in the 3’ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol 79(Pt 4):715–718
DuBois RM et al (2013) Crystal structure of the avian astrovirus capsid spike. J Virol 87(14):7853–7863
Toh Y et al (2016) Crystal structure of the human astrovirus capsid protein. J Virol 90(20):9008–9017
Smyth VJ et al (2012) Capsid protein sequence diversity of chicken astrovirus. Avian Pathol 41(2):151–159
Jonassen CM et al (2001) Comparison of capsid sequences from human and animal astroviruses. J Gen Virol 82(Pt 5):1061–1067
Dong J et al (2011) Crystal structure of the human astrovirus capsid spike. Proc Natl Acad Sci U S A 108(31):12681–12686
Funding
This work was supported by the key underprop project of Tianjin Municipal Science and Technology Bureau (20YFZCSN00340) and the Tianjin Synthetic biotechnology innovation capability improvement project in China (TSBICIP-KJGG-014).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: JH H. Performed the experiments: Z T, H Z, RQ S, RY X, LL Z, and ZX L. Analyzed the data: H Z and JH H. Contributed reagents/materials /analysis tools: JH H. Wrote the paper: ZX L and JH H.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Luis Nero
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tan, Z., Zhai, H., Sun, R. et al. Complete genome sequence and phylogenetic analysis of a goose astrovirus isolate in China. Braz J Microbiol 54, 427–434 (2023). https://doi.org/10.1007/s42770-022-00854-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-022-00854-7