Abstract
The heterogeneous character of fixed-film reactors may create highly specialized zones with a stratified distribution of microbial groups and varying capabilities to withstand high organic loads in anaerobic digestion (AD) systems. The microbial distribution and methane-producing potential of biomass from different regions (feeding zone and structured bed) of two second-stage thermophilic (55 ºC) fixed-film reactors were assessed. Three levels of food-to-microorganism (F/M) ratio (0.4, 1.0 and 3.0 g-COD g−1VS) using fermented (two-stage AD) and fresh (single-stage AD) sugarcane molasses were tested in batch reactors, simulating low to high organic loads. Specific methane production rates increased as the F/M increased when using fermented molasses, maintaining efficient methanogenesis at substrate availability levels threefold higher than single-stage schemes (3.0 vs. 1.0 g-COD g−1VS). Success in methane production derived from the homogenous establishment (similar in both feeding zone and bed) of syntrophic associations between acetogens (Pelotomaculum, Syntrophothermus, Syntrophomonas and Thermodesulfovibrio), acetate oxidizers (Thermoacetogenium, Mesotoga and Pseudothermotoga) and hydrogenotrophic methogens (Methanothermobacter and Methanoculleus) replacing acetoclastic methanogens (Methanosaeta). Phase separation under thermophilic conditions was demonstrated to boost methane production from sugar-rich substrates, because the process depends on microbial groups (hydrogenotrophs) that grow faster and are less susceptible to low pH values compared to acetotrophs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anaerobic digestion (AD) of high-strength wastewaters has been facilitated by developing high-rate systems, in which biomass retention mechanisms enabled uncoupling the hydraulic retention time (HRT) from the solid retention time in reactors (van Lier et al. 2015). Biomass granulation or the provision of stationary or moving media for cell attachment has created suitable conditions for the application of high organic loading rate (OLR) under low HRT, resulting in a series of benefits, such as achieving higher robustness and operating stability, requiring more compact units and increasing biogas production. Among the numerous configurations proposed are two that successfully achieved effective full-scale applications, namely, the sludge blanket and fixed-film systems (Moletta 2005). The vulnerability of sludge granulation to compositional characteristics commonly found in numerous wastewaters, such as high suspended solid content and high salinity (van Lier et al. 2015), provides some advantages to using support materials in cell retention. Focus is given to the second group in this study, because recent research efforts have led to improvements on the conventional packed-bed systems through vertically arranging the support media to characterize the anaerobic structured-bed reactor (AnSTBR) (Aquino et al. 2017; Camiloti et al. 2014; Fuess et al. 2021a; Mockaitis et al. 2014).
AnSTBR systems simultaneously provide high void indices and adequate surface area levels for biomass attachment, which expand the applicability of fixed-film reactors. Compared to packed-bed systems, the higher bed porosity (> 90%) (Aquino et al. 2017; Mockaitis et al. 2014) minimizes bed clogging-related limitations, which enables both the processing of solid-rich substrates and the establishment of microbial populations with high growth yields (e.g. hydrolytic and fermentative bacteria). The attached growth itself also comprises an advantageous feature compared to other high-rate systems because less metabolic energy is required to establish high cell densities within the system (Aquino et al. 2017; Blanco et al. 2017). Consequently, the microbial community presents higher robustness when faced with adverse environmental conditions, such as organic overloading events (Chan et al. 2009).
Interestingly, fixed-film reactors do not behave as homogeneous systems, because the low-to-null cell mobility associated with the progressive conversion of the substrate may create highly specialized zones in the reactor. Following the flow direction, a “more acidogenic” microbial community should be expected in regions with high fresh substrate availability (e.g. feeding zones), whilst a “more acetogenic” and mainly methanogenic-like character should be observed in the subsequent compartments of the reactor. It is worth highlighting that the fermentability of the substrate may directly impact this microbial distribution, because the proposed specialization should be more evident in the processing of easily fermentable substrates, such as sugar-rich ones. Conversely, a relatively more homogeneous microbial distribution (or at least without the prevalence of fermentative groups near the feeding zone) is expected to occur in second-stage methanogenic reactors, in AD schemes where acidogenesis is carried out separately from methanogenesis.
Different microbial populations will have different growth rates (Mosey 1983), potentially leading to wide variations of the food-to-microorganism (F/M) ratio along fixed-film reactors. The amount of microorganisms is commonly measured as volatile suspended solids (VSS) or only volatile solids (VS), whilst the “food” is represented by the organic matter content measured as the chemical oxygen demand (COD). Recent studies associated optimal fermentation activity (targeting biohydrogen production) with F/M ratios within the range of 4.0–6.0 g-COD g−1VSS d−1 (Blanco et al. 2017; Anzola-Rojas et al. 2015; Fuess et al. 2021b; Hafez et al. 2010), whilst stable methanogenesis has been conventionally pointed to occur under much lower F/M ratio values (< 0.4 g-COD g−1VSS d−1 (Aquino et al. 2017; Barros et al. 2016; Chernicharo 2007), specifically in single-stage schemes. However, the compartmentalized assessment of a second-stage AnSTBR indicated the establishment of highly efficient substrate conversion into methane under F/M ratios as high as 3.0 g-COD g−1VSS d−1 (Fuess et al., 2021a) directly contradicting the expected behavior for methanogenic environments. In practical aspects, high F/M ratios indicate (or simulate, in the case of batch reactors) situations of organic overloading (or close to that), in which the activity of fermentative bacteria should prevail and inhibit methanogens by providing an excess amount of volatile fatty acids (VFA). Synonyms for the term F/M ratio in continuous reactors include both “biological loading rate” and “sludge loading rate” (Chernicharo 2007), as well as “specific organic loading rate” (Anzola-Rojas et al. 2015).
Some important questions arise from the stratified character of fixed-film reactors, regarding both the spatial distribution of specialized microbial groups and variations in the quality of the substrate (driven by the pre-fermentation intensity): [i] Does biomass specialization effectively occur as a result of differences in substrate quality (following the flow direction) or is a homogeneous distribution of all microbial groups participating in substrate conversion repeated along the entire reactor? [ii] More specifically, can methanogens withstand higher organic loads (or higher F/M ratios) when phase separation is used, i.e. when substrate fermentation is carried out (separately) prior to methanogenesis?
To answer these questions, the microbial distribution and methane-producing potential of biomass collected in different regions of two second-stage thermophilic AnSTBR systems were assessed. Samples collected from the feeding zone and the bed region were compared using 16S rRNA gene sequencing to understand performance discrepancies as a function of the prevailing microbial groups. Batch tests using biomass from the feeding zones were further conducted under different F/M ratio values using fermented and fresh (non-fermented) sugarcane molasses as the substrate to understand how efficiently methanogenic consortia are able to perform under low and excess substrate availability levels, potentially defining operating limits for anaerobic reactors.
Material and methods
AnSTBR systems and thermophilic methanogenic consortia
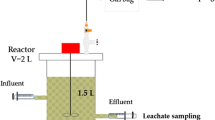
Two bench-scale (1.9 L each) AnSTBR systems were sampled to obtain the methanogenic consortia microbiologically characterized and used as inocula in methane production potential (MPP) tests in this study. Polyurethane (PU) foam strips wrapped in spiral polyvinyl chloride (PVC) frames were used as the support material in both reactors, which were operated in upflow mode and fed with fermented sugarcane molasses for 250 (RM1) and 230 (RM2) days under thermophilic temperature conditions (55 ºC). The reactors were subjected to different alkalinization strategies: while sodium bicarbonate (NaHCO3) dosing was carried out during the entire operating period of RM1, sodium hydroxide (NaOH) dosing coupled to effluent recirculation was initially used in RM2 (185 d), after which this approach was replaced by NaHCO3 dosing. Increasing OLR levels were applied to both systems to define the operating limit conditions for each alkalinization strategy. Complete details of the long-term operation of RM1 and RM2 were previously presented elsewhere (Fuess et al. 2021a). Despite the differences aforementioned, the operation of both reactors was finalized under equivalent conditions of OLR (10.0 kg-CODt m−3 d−1; CODt = total chemical oxygen demand), HRT (24.0 h) and influent CODt (10 g L−1), in addition to NaHCO3 dosing. Biomass samples were further collected during reactor disassembling separately from the feeding zone (suspended cells in the bulk liquid) and from the bed region (PU strips—attached cells). Attached biomass was removed from the media by washing the strips with distilled water. Sample cleansing prior to DNA extraction was carried out immediately after collection, whilst the remaining amounts of biomass from the feeding zones were stored at 4 ºC prior to use in MPP tests. Figure 1 shows constructive aspects of the AnSTBR and details of the biomass collection during the disassembling procedure.
Details of the AnSTBR systems (simplified sketch and experimental prior to inoculation) and biomass sampling from the feeding zone (FDZ) and structured bed (STB) during reactor disassembling. The same procedure was applied in RM1 and RM2. Legend: 1- reactor feeding, 2- effluent collection, 3- biogas collection
Sugarcane molasses characterization
Fresh sugarcane molasses (Mol) was collected from a large scale annexed biorefinery (milling approximately 10 million tons of sugarcane per harvest) located in Pradópolis, São Paulo, Brazil. The samples were stored at 4 ºC prior to use in both the feeding of the acidogenic reactor (see further description) and MPP tests. Fermented molasses (fMol) was collected from a bench-scale (2.0 L) thermophilic (55 ºC) acidogenic AnSTBR operated under optimized conditions (OLR = 86.0 kg-CODt m−3 d−1; HRT = 10.0 h) targeting enhanced biohydrogen production, as reported elsewhere (Fuess et al. 2021b). fMol samples were stored at −20 ºC prior to use (feeding both RM1 and RM2 and as substrate in MPP tests) to interrupt the activity of cells washed-out from the acidogenic reactor and preserve the compositional aspects of the fermented substrate. Detailed compositional aspects of Mol and fMol are presented in Table 1.
Methane production potential tests
MPP tests were carried out in 500 mL (nominal volume) Erlenmeyer flasks (with DIN standard PBT screw caps—GL45) filled with 100 mL of substrate and inoculum. Both Mol and fMol were diluted to obtain a COD of 10 g L−1, simulating the terminal operating condition applied in the continuous reactors. Sugars were promptly solubilized once Mol was diluted. Meanwhile, fMol was centrifuged (9000 rpm, 5 min) prior to dilution as a strategy to remove cells (fermentative biomass) remaining from the acidogenic reactors, i.e. only the supernatant was used. In practical aspects, only the soluble organic content of both Mol and fMol was used as the substrate; however, the nomenclature “soluble COD (CODs)” was not used in this specific step because no 0.45 μm-filtration was carried out. Conversely, methanogenic sludge sources were centrifuged under similar conditions (9000 rpm, 10 min) to remove excess liquid phase, i.e. only the pellets were used, which were called wet sludge. In addition to biomass samples collected from the feeding zones of RM1 and RM2, the thermophilic methanogenic sludge used in the inoculation of both continuous reactors (identified as IN) was also assessed in MPP tests, following the same pre-centrifugation step. Assessing only biomass samples from the feeding zone was based on the performance of the continuous reactors, because the CODs was majorly consumed (> 70%) in this compartment in both reactors (Fuess et al. 2021a).
Once the volatile solids (VS) content was measured in each methanogenic consortium, the amount of wet sludge added to each flask was calculated according to three different F/M (or substrate-to-inoculum) ratios, namely, 0.4, 1.0 and 3.0 g-COD g−1VS. The selected F/M ratios were based on the specific organic loading rate (sOLR) assessed in the continuous reactors, specifically in RM1 (Fuess et al. 2021a): the values 0.4 and 1.0 g-COD g−1VS simulated the sOLR (obtained in terms of g-COD g−1VSS d−1) observed under stable operating conditions, using the total amount of biomass inside the reactor and the amount of biomass retained specifically in the feeding zone as the references, respectively. In turn, the value of 3.0 g-COD g−1VS represented the maximum sOLR observed in the feeding chamber still associated with a stable operation. The three levels of the F/M ratio were assessed in fMol-fed reactors using all sludge sources (RM1, RM2 and IN). In the particular case of Mol, only sludge from RM2 was used. Finally, blank reactors were also monitored (duplicates) by replacing Mol or fMol with distilled water, using a fixed mass of 10 g of wet sludge per flask. For identification purposes, the nomenclature used to differentiate the conditions was formed by three parts: F/M ratio value-inoculum source(substrate type)—e.g. 0.4-RM1(fMol) and 3.0-RM2(Mol). The adopted experimental arrangement enabled comparing two- and single-stage AD schemes.
In addition to the definitions regarding both the F/M ratio and inoculum sources, reactor alkalinization (including blanks) was based on NaHCO3 dosing (0.25 g-NaHCO3 g−1COD), also using the continuous reactors as the reference (Fuess et al. 2021a). pH values in fMol and Mol after NaHCO3 dosing reached ca. 6.70 and 7.60, respectively. After filling the flasks with alkalinized substrate and sludge, the reactors were fluxed with ultra-high purity grade nitrogen for 5 min to establish conditions of anaerobiosis and sealed with rubber stoppers and plastic caps. The reactors were incubated in an orbital shaker (model Multitron PRO Incubator Shaker—Infors HT, Infors AG, Bottmingen-Basel, Switzerland) and monitored for periods ranging between 7 and 16 d, depending mainly on the temporal evolution of methane production. Temperature and agitation conditions were set respectively as 55 ºC and 100 rpm.
Reactor monitoring was carried out through periodically sampling both the gas and liquid phases with 1 mL insulin syringes. In the specific case of biogas collection, the syringe was equipped with a Teflon body two-way valve (Supelco™ Analytical—Sigma Aldrich, Bellefonte, PA, USA). The sampling periodicity varied according to the response of the systems in terms of the cumulative methane production and COD decay. Experimental runs were finalized after observing coefficient of variation (CV) values lower than 5% for at least three consecutive points of the cumulative methane production, whilst the stabilization of COD decay was used as a secondary reference. Liquid phase sampling was limited to 10% of the total substrate volume (10 mL). Biogas production was monitored by measuring the internal pressure of the flasks followed by the compositional analysis of the sample. Pressure measurements were carried out using a pressure gauge model TPR-16 (Desin Instruments, s.a., Barcelona, Spain), with an upper measurement limit of 500 mbar (50 kPa). Whenever required, pressure reliefs were conducted to prevent the occurrence of pressure values out of the measurement range.
Analytical methods, calculations and kinetic analysis
Overall liquid phase measurements included determining the pH, CODt, CODs, total carbohydrates (CHt), lactate, total phenols (PheOH), partial (PA) and intermediate (IA) alkalinity, volatile organic acids by titration (VOAtit), volatile organic acids (VOAGC; C2–C6) and solvents by gas chromatography (GC) and sulfate (SO42−). The periodic monitoring of the liquid phase in MPP tests included exclusively CODs measurements, whilst all remaining parameters were measured in non-diluted substrates (Table 1; except for PA, IA and VOAtit) and by the end of the incubation periods (except for the CODt). COD (both total and soluble), pH and SO42− measurements were based on the Standard Methods for the Examination of Water and Wastewater (APHA et al. 2012), whilst specific protocols were used for the remaining parameters: CHt (Dubois et al. 1956), lactate (Taylor 1996), PheOH (Buchanan and Nicell 1997), PA/IA (Ripley et al. 1986), VOAtit (Kapp 1984) and VOAGC/solvents (Adorno et al. 2014). Except for the case of CODt determination, samples were filtered in 0.45 μm syringe membranes (Chromafil GF/PET, Macherey–Nagel GmbH & Co. KG, Düren, Germany) prior to all remaining analyses.
Considering gas phase monitoring, biogas composition (nitrogen—N2, methane—CH4, carbon dioxide—CO2 and hydrogen sulfide—H2S) measurements were carried out in a GC equipped with a thermal conductivity detector (model GC-2014, Shimadzu Scientific Instruments, Japan) using hydrogen as the carrier gas (Lebrero et al. 2016). Specifically, the cumulative methane production up to time “n” (VCH4,n; NmL) was calculated as proposed elsewhere (Santos et al. 2019) using Eq. (1), in which the terms iPpre-inj,n, iPpost-inj,n-1, fCH4,n, Vheadspace, T0, P0 and Texp, are, respectively, the internal pressure prior to sampling at time “n” (atm), the internal pressure after sampling at time “n—1” (atm), the methane fraction in biogas at time “n” (dimensionless), the volume of the headspace (L), the temperature at normal conditions (273.15 K), the pressure at normal conditions (1 atm) and the incubation temperature (55 ºC = 328.15 K).
The modified Gompertz model (Zwietering et al. 1990) (Eq. 2) was fitted to VCH4 values to obtain the potential methane production (PCH4; NmL), the maximum methane production rate (RCH4; NmL h−1) and the lag phase period (λ; h). The maximum methane production rate was also calculated in specific terms (RCH4’; NmL h−1 g−1VS) by dividing RCH4 by the amount of sludge (VS) used in the inoculation of each reactor. In Eq. (2), the terms VCH4(t) and e are, respectively, the cumulative methane production (model) as a function of the incubation period (t) and the Euler’s number (2.71828). Endogenous methane production (assessed in blank reactors) was discounted from methane production values obtained when processing fMol and Mol. Hence, VCH4 values (both experimental and fitted) correspond to the net methane production. In parallel, kinetics of substrate consumption (CODs) was assessed through fitting the first-order decay model with residual (Rodrigues et al. 2003) (Eq. 3) to CODs temporal profiles, from which the residual CODs (CODsR; mg L−1), the initial CODs (CODs0; mg L−1) and the first-order kinetic constant (k1; h−1) were estimated. The specific first-order kinetic constant (k1’; h−1 g−1VS) was calculated analogously to RCH4’. Model fitting was carried out using the software Origin 2020 (OriginLab Corporation, Northampton, MA, USA) using the Levenberg–Marquardt algorithm. In the particular case of methane production, models were adjusted to experimental points collected up to fulfilling two criteria: [i] cumulative CODs removal efficiency (ERCODs; Eq. 4) higher than 70% and [ii] CODs removal efficiency between two consecutive points (ERCODs,interval; Eq. 5) lower than 5%. In Eq. (4)–(5) the terms CODsn, CODs0 and CODsn-1 are the CODs (mg L−1) measured at times “n”, 0 (zero) and “n – 1”, respectively.
Microbial community characterization by 16S rRNA gene sequencing
Biomass sampling and sample preparation.
Preliminary details of the microbial community sampling (targeting microbial characterization) were presented in Section “AnSTBR systems and thermophilic methanogenic consortia”. Samples from suspended cells in the feeding zone and attached cells from the bed region were obtained for both continuous methanogenic reactors (Fig. 1) during reactor disassembling and promptly stored at – 20 ºC after cleansing, as described in the sequence. The thermophilic sludge (IN) used in the inoculation of RM1 and RM2 (collected from a thermophilic full-scale UASB processing sugarcane vinasse) was also characterized. Because the COVID-19 pandemic forced a relatively prolonged storage (4 ºC) of the remaining biomass samples from the feeding zones (ca. 6 months) and from IN (ca. 14 months) prior to MPP tests, aliquots of the stored samples were also analyzed (except for the case of RM1). Hence, a total of seven samples were processed and were identified by the sludge source (IN, RM1 or RM2) and the reactor region, i.e. feeding zone (FDZ) or structured bed (STB). The nomenclature “ps” (post-storage) was also used when pertinent. Table 2 brings details of the timeline followed to obtain biomass samples. All samples were subjected to 2–3 rounds of centrifugation (6000 rpm, 10 min) using a phosphate-buffered saline, which aimed to remove impurities (residual organic compounds). Finally, biomass pallets were stored at – 20 ºC prior to further processing.
DNA extraction, 16S rRNA gene amplicon sequencing and bioinformatics
The procedures used in DNA extraction and 16S rRNA gene sequencing are presented elsewhere (Piffer et al. 2022)—refer to Supplementary material for details. Sequences were analyzed using quantitative insights into a microbial ecology’ pipeline (QIIME2 2021.4 release) (Bolyen et al. 2019). Demultiplexed paired-end sequencing reads were imported into the QIIME2, resulting in 358,837 joined sequences. Sequencing reads were ‘denoised’ using the ‘divisive amplicon denoising algorithm’ DADA2 (Callahan et al. 2016) plugin in QIIME2, which provided high resolution amplicon sequence variants (ASV) for downstream analysis by filtering out noises, correcting errors in marginal sequences, removing chimeric sequences and singletons and dereplicating the resulting sequences. As a result, 313,539 sequences were obtained, ranging from 39,937 to 46,692 per sample (n = 7 in total), representing 719 ASV. The consensus sequences for the ASV were classified with a classify-sklearn classifier trained against the most recent SILVA 16S rRNA gene reference (release 138.1) database (Quast et al. 2013). In order to complete downstream diversity and composition analyses, sequences were rarefied to the lowest number of sequences per sample (n = 39,937 sequences). The sequences were submitted to the National Center for Biotechnology Information (NCBI; http://ncbi.nlm.nih.gov) under accession BioProject ID PRJNA778433.
Data analysis and visualization
All the statistical analyses were performed in R version 3.5.1 with R Studio environment, Version 1.3.1093. The biom file from QIIME2 was imported and analyzed through phyloseq-modified workflow (McMurdie and Holmes 2013). Taxon relative abundance bar charts were generated using custom R scripts and ggplot2 (v3.3.2) (Wickham 2016). A similarity matrix of weighted UniFrac distances was used for ordination by principal coordinates analysis (PCoA) of non-transformed relative abundance diversity datasets implemented in phyloseq and plotted using ggplot2 to visualize the distribution of the microbial composition. Heat maps were generated using ampvis2 (v.2.6.5) (Andersen et al. 2018).
Results and discussion
Kinetics of methane production
The methane production potential of thermophilic microbial consortia processing sugarcane molasses was assessed under different F/M ratios, simulating different levels of substrate availability and also comparing single- and two-stage AD schemes. Temporal profiles for the cumulative methane production and CODs decay (experimental data) are depicted in Figs. 2 and 3, respectively. The kinetic parameters of the fitted models describing methane production and CODs decay are detailed in Tables 3 and 4, respectively. RCH4 values followed an increasing pattern as the F/M ratio decreased (Table 3), regardless of the inoculum source and substrate type. This pattern characterizes an expected result, because higher biomass amounts (lower F/M ratios) will have the ability to promptly convert the available substrate. However, an inverse pattern was observed when considering RCH4’ values, which increased as the F/M ratio was increased for a given inoculum source (Table 3; considering specifically the use of fMol). This result indicates that lower amounts of biomass produced methane more efficiently, most likely due to the favoring of substrate conversion kinetics under excess substrate availability (prior to reaching the limit concentration, i.e. CODs, prior to inhibition). Under conditions of excess biomass (lower F/M ratios), a fraction of the cell most likely remained in a latency-like state, without effectively participating in methane evolution. From a kinetic perspective, lower substrate availability was the limiting factor for methanogens.
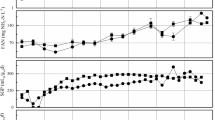
An integrated analysis including RCH4’ values obtained for RM1-derived sludge (this study) and specific methane production rate values in the different operating phases of RM1 (Fuess et al. 2021a) revealed a consistent variation pattern for RCH4’ (Fig. 4a–b), regardless of the operating mode, i.e. batch or continuous. The specific methane production rate in RM1 (at the different OLR levels) was calculated through re-arranging performance data previously presented, namely, the volumetric methane production rate and the biomass retention, providing a novel response not addressed in the base reference study (Fuess et al. 2021a). Details of this calculation are presented in the Supplementary data section. Despite the different measurement units for the F/M ratio (g-COD g−1VS) and the sOLR (g-COD g−1VSS d−1), both parameters measure the amount of substrate available for conversion in the reactors on an equivalent basis, which supports the integrated comparison. RCH4’ varied linearly up to the maximum experimental sOLR measured in RM1 (~ 5.0 g-COD g−1VSS d−1; Fig. 4a), indicating that F/M ratio values assessed in batch tests (≤ 3.0 g-COD g−1VS) characterized conditions of excess sludge compared to the amount of substrate, justifying the increasing trend previously highlighted. Differently from batch systems, in which the initial F/M is defined and fixed to a desired value, the sOLR varies as a function of both the applied OLR and the amount of retained biomass within continuous reactors (Fuess et al. 2021a; Anzola-Rojas et al. 2015), explaining the varying values in RM1. Interestingly, specific methane production was subjected to some degree of inhibition with the increase in the OLR (Fig. 4b), but not as a direct consequence of excess substrate availability. With the continuous retention of biomass within the continuous system, increasing the OLR (up to 20 kg-COD m−3 d−1) was not sufficient to proportionally increase the sOLR (Fig. 4b), explaining the lower RCH4’ values. Hence, the F/M ratios assessed in the batch tests may represent two operating situations of the continuous reactors: a short-term operation characterized by a low OLR under equally low biomass retention levels or a long-term operation with relatively high OLR and excess sludge retention.
Correlations between methane production and substrate availability: a maximum specific methane production rate (RCH4’) as function of the F/M ratio (batch reactors) and specific organic loading rate (sOLR, continuous reactor) and b RCH4’ as function of the organic loading rate (OLR) and sOLR (continuous reactor). Note: aValues calculated through re-arranging performance data collected from RM1 presented elsewhere (Fuess et al. 2021a)
An inverse pattern was observed when using fresh molasses as the substrate, in which lower RCH4’ values were observed at higher F/M ratios (Table 3), until reaching a marked inhibition of the methanogenic activity in the highest F/M ratio level (3.0 g-COD g−1VS) assessed (Fig. 2c). The carbohydrate-rich composition of Mol (> 80% of the CODs; Table 1) promptly favored the growth of fermentative bacteria, diverting the electron flow, i.e. CODs consumption, from methanogenesis towards cell synthesis. Similar results were observed in previous studies on the biodigestion of highly fermentable substrates, such as sugarcane juice and molasses (Fuess et al. 2021c; Vilela et al. 2021) and glycerol-supplemented sugarcane vinasse (Borges et al. 2022). In all cases, the impairment of the methanogenic activity was not impeditive for achieving relatively high substrate consumption levels, reinforcing the diversion towards cell synthesis. Comparing methane evolution patterns (and the kinetic parameters estimated) for the RM2(fMol) and RM2(Mol) conditions (Fig. 2 and Table 3) supports this hypothesis, because the lowest RCH4 and RCH4’ values were always associated with the non-fermented molasses (for a given F/M ratio). In practical aspects, this discrepancy demonstrated that phase separation increases the capability of anaerobic systems to withstand organic overloading conditions (simulated by high F/M ratios) far beyond the limits achieved in single-stage AD. The experimental arrangement indicated that two-stage schemes withstand at least a threefold higher organic load compared to single-stage ones, because methane production collapsed in condition 3.0-RM2(Mol)—despite the observation of methane production in this case (Fig. 2c), terminal pH values lower than 5.0 were observed, i.e. an evidence of VFA buildup. Hence, starting up a second-stage methanogenic reactor should require lower amounts of sludge and would demand a shorter period compared to single-stage schemes.
Shifting the analysis perspective, the comparison between the different biomass sources assessed (in a fixed F/M ratio) indicated that RCH4/RCH4’ values always followed the order RM1 > RM2 > IN (Table 3), suggesting that biomass collected in RM1 was better adapted to the substrate and operation (incubation) conditions than the other inocula. Regardless of the differences observed in the long-term performance of RM1 and RM2, as a direct consequence of different alkalinization strategies, CODs removal efficiency in the feeding zone of both reactors was equivalent, i.e. 74.9% (RM1) and 71.8% (RM2) (Fuess et al. 2021a), indicating that equivalent methane evolution patterns should be observed in MPP tests. Differences in the prevailing microbial communities (also resulting from the different alkalinization approaches) may explain the observed patterns (Section “Microbial community composition”). It is worth highlighting the significant discrepancy in the performance of methanogenesis of IN relative to RM1/RM2, mainly with respect to much longer lag phase periods (Table 3), which resulted from the continuous adaptation of the thermophilic biomass to fermented molasses in RM1 and RM2 for periods of at least 230 d. Besides the microbial differences (Section “Microbial community composition”), the OLR applied in the UASB from which IN was collected (~ 5.0 kg-CODt m−3 d−1) is half of that applied in RM1 and RM2 (10 kg-CODt m−3 d−1) when microbial community samples were obtained, indicating a higher capability to withstand high substrate availability levels in the latter. However, the non-adaptation of the biomass to the incubation conditions was attenuated at high substrate availability: when comparing the different inocula (fMol-fed reactors), the higher the F/M ratio, the lower the variation coefficient calculated for RCH4 (or RCH4’) values, i.e. 30%, 40% and 50% respectively for the F/M ratios of 3.0, 1.0, 0.4 g-COD g−1VS.
Kinetics of substrate consumption
Differently from methane production, CODs decay followed distinct kinetic patterns, according to both the sludge source and F/M ratio (Fig. 3 and Table 4). Single kinetic decays were observed only when assessing an F/M ratio of 0.4 g-COD g−1VS in biomass samples from RM1 and RM2 (considering both fMol and Mol in the latter). The relative excess sludge amount in such cases favored a prompt substrate consumption, with a wide advantage for the case of RM1 (k1’ = 1.384 × 10–2 h−1 g−1VS; Table 4). Single decay patterns were also observed in the case of IN (F/M ratios of 0.4 and 1.0 g-COD g−1VS); however, only after 106 h of experimental run (Fig. 3a-b), suggesting a progressive adaptation of the inoculum to the incubation conditions. In all remaining cases, substrate decay followed two distinct patterns, either with an initial linear decay (condition 3.0-RM1(fMol)) or with two sequential exponential decays (conditions 1.0-RM2(fMol), 3.0-RM2(fMol), 3.0-IN(fMol) and 1.0-RM2(Mol)) (Table 4). In these conditions, no trend repetition was observed, i.e. while k1’ in the first decay period exceed that of the second in 1.0-RM2(fMol) and 3.0-IN(fMol), the opposite was observed in 3.0-RM2(fMol) and 1.0-RM2(Mol).
For a given sludge source (fed with fMol), the higher the F/M ratio, the higher the value of k1’ (Table 4), repeating the behavior previously observed and described for RCH4’ (Table 3). Complementing the hypothesis of higher metabolic efficiency towards methane production, substrate consumption also includes processes other than methanogenesis, such as cell synthesis. Hence, the high k1’ values estimated under lower biomass amounts (high F/M ratios) most likely resulted from both metabolic steps. In fact, relatively high biomass concentrations, i.e. M > F, are recommended when assessing the methanogenic activity of anaerobic sludge sources to minimize the negative effects of excess biomass synthesis over methane production (Aquino et al. 2007; Holliger et al. 2016). The establishment of enhanced cell growth as a significant electron (substrate) sink also justifies the CODs consumption under unfavorable conditions to methanogenesis when using fresh molasses, mainly under high F/M ratios, i.e. 1.0 and 3.0 g-COD g−1VS. These values are much higher than the ones previously recommended for efficient methane production from easily fermentable substrates, i.e. inoculum-to-substrate ratio higher than 4.0 (or F/M ratio lower than 0.25) (Holliger et al. 2016). Revisiting the parallel with reactor startup, efficient substrate conversion rates coupled to short latency periods will only be achieved when sufficient amounts of biomass are available (Haider et al. 2015). Sulfate reduction could have also contributed to the CODs decay, because sulfidogenesis can be associated with the complete oxidation of organic compounds. However, a significant deviation of electrons towards sulfate reduction should only be observed at high sulfate availability (COD/sulfate < 25.0) (Kiyuna et al. 2017), which was not observed for molasses, regardless of the pre-fermentation (COD/sulfate > 60.0; Table 1).
Similarly to the case of RCH4’, k1’ values followed the same order for a given F/M ratio, i.e. RM1 > RM2 > IN (Table 4), so that the discrepancy may be understood from an analogous perspective, focusing on microbial characterization aspects (Section “Microbial community composition”). It is worth highlighting that enhanced CODs decay in Mol-fed reactors at lower F/M ratios (≤ 1.0 g-COD g−1VS) was anticipated in comparison with IN (Fig. 3a-b), despite using fermented molasses in the latter. A similar response occurred in the case of methane evolution from fresh molasses at the lowest F/M ratio (0.4 g-COD g−1VS; Fig. 2a), in which the exponential production phase anticipated the one from fermented molasses in reactors inoculated with IN. These results may be explained by the long-term (≥ 230 d) adaptation of IN in the continuous reactors, which attenuated negative effects of enhanced VFA production over methanogens when processing Mol under higher sludge concentrations. Hence, the efficacy of phase separation in high-rate AD systems cannot be assessed separately from the degree of adaptation of the sludge to the desired operating conditions.
Breakdown of liquid phase constituents in biodigested molasses
Details of the liquid phase constituents at the end of the incubation periods are depicted in Fig. 5, including the terminal pH, the balance between bicarbonate alkalinity and VFA accumulation and the breakdown of the CODs. Different terminal pH values were observed according to the type of substrate, i.e. approximately 8.0 and 7.5 for fermented (regardless of the F/M ratio) and fresh (except for the F/M ratio of 3.0 g-COD g−1VS) molasses (Fig. 5a). While a marked increase relative to the pH of fermented molasses supplied with NaHCO3 prior to incubation (< 7.0) was observed in fMol-fed reactors, initial and terminal pH values were equivalent (~ 7.5) when using fresh molasses (F/M ratios of 0.4 and 1.0 g-COD g−1VS). In the first case, the non-occurrence of enhanced sugar fermentation positively impacted the balance of partial (bicarbonate) alkalinity (Fig. 5b), with values 1.5-to-2.0-fold higher than in reactors fed with fresh molasses (considering equivalent F/M ratios). These patterns (associated with the higher terminal pH values) indicate an excess of NaHCO3 supply in second-stage methanogenic systems, suggesting the possibility to use lower doses without impairing the performance of methanogenesis. In fact, dosing 0.20 g-NaHCO3 g−1COD in RM1 (continuous reactor) did not negatively impact system performance (Fuess et al. 2021a). Conversely, removing NaHCO3 supply triggered the inhibition of methanogenesis in a continuous reactor (also an AnSTBR) fed with fresh molasses, so that a minimum dose of 0.25 g-NaHCO3 g−1COD was defined as the limiting condition to maintain stable performance levels (Oliveira et al. 2020). As a side-effect remark of the experiment, dosing equivalent amounts of NaHCO3 (0.25 g g−1COD) in methanogenic systems fed with fresh and fermented substrates results in excess bicarbonate alkalinity in the latter (up to 200% higher concentrations than in the first). In practical aspects, NaHCO3 dosing in two-stage biodigestion systems have potential to be decreased to economically cost-competitive levels, improving mechanisms to ensure the maintenance of operating stability in industrial scale plants.
Characterization of biodigested molasses at the end of the incubation periods: a pH,b partial alkalinity (PA), c intermediate-to-partial alkalinity (IA/PA) ratio, d concentration of volatile organic acids by titration (VOAtit), e breakdown of VFA (using gas chromatography) and f breakdown of the CODs. Legend: aAfter NaHCO3 dosing, (*) indicates null values
The intermediate-to-partial alkalinity (IA/PA) ratio measured at the end of the incubation periods reached values lower than 0.16 in all cases in which methane production was not inhibited (Fig. 5c), indicating much higher participation of bicarbonate-derived alkalinity than the one related to organic acids (despite the aforementioned discrepant patterns). Overall, the lower the F/M ratio, the lower the IA/PA value for a given source of inoculum (Fig. 5c), characterizing a relative accumulation of organic acids at high F/M ratios (VOAtit usually below 500 mg L−1; Fig. 5d). Residual VOAtit concentrations measured in reactors inoculated with biomass from RM1 (426 and 718 mg L−1 in F/M ratios of 1.0 and 3.0 g-COD g−1VS, respectively; Fig. 5d) were higher than those in reactors using RM2-derived sludge (< 283 mg L−1; Fig. 5d) most likely due to differences in the incubation periods. While methane production (CV < 5%; Section “Methane production potential tests”) and the associated CODs decay met the criteria for finalizing the experimental runs within 163 h in the case of RM1, reactors inoculated with sludge from RM2 required longer periods, which varied from 192 h (F/M = 0.4 and 1.0 g-COD g−1VS) to 216 h (F/M = 3.0 g-COD g−1VS). Enhanced acidification (VOAtit = 4500 mg L−1; Fig. 5d) was only observed in condition 3.0-RM2(Mol), in which the inhibition of methanogenesis (Fig. 2c) was associated with terminal pH values lower than 5.0 (Fig. 5a), as well as with the full consumption of the partial alkalinity (Fig. 5b).
The distribution of residual VFA concentrations (Fig. 5e) indicated distinct patterns according to the success in the establishment of methanogenesis. Propionate was the prevailing metabolite in conditions in which methane production evolved over time, regardless of differences in the kinetic patterns and type of substrate (Fig. 5e). In these cases, propionate oxidation into acetate and the subsequent oxidation of acetate into CO2 and hydrogen most likely characterized the limiting steps, slowing down methane evolution via the hydrogenotrophic pathway, i.e. the primary methanogenic pathway established in all reactors, as supported by the microbial characterization of the inocula (Section “Microbial community composition”). Differently, acetate buildup was observed in condition 3.0-RM2(Mol), i.e. the only case in which methane production was impaired (Fig. 5e). In addition to the failure in acetate oxidation, which may have also limited the activity of hydrogenotrophic methanogens in this case, the activity of acetoclastic methanogens was equally inefficient to convert the excess acetate available, most likely due to the inhibition by the low pH (Hao et al. 2012).
Apart from the accumulation of VFA, the breakdown of the terminal CODs indicated consistent participation of total phenols (PheOH) in the group of non-converted organic matter, with most values within the range of 20 to 40% (Fig. 5f). PheOH usually accounts for a significant fraction of the residual COD in biodigested sugarcane-derived substrates, such as vinasse (Santos et al. 2019) and molasses (Fuess et al. 2021a). The presence of highly recalcitrant melanoidins (from the Maillard reaction between sugars and proteins) and colorants (except caramels) configures the main natural sources of phenols in these substrates (Chandra et al. 2018; Mohana et al. 2009), providing their characteristic dark brown color. It is noteworthy that the participation of PheOH in the CODs increased in all assessed conditions, i.e. from 2.1–2.7% (non-biodigested substrates; Table 1) to (6.2–40.5% (biodigested substrates; Fig. 5e), with the highest terminal values usually observed at excess biomass concentrations (F/M = 0.4 and 1.0 g-COD g−1VS). This pattern most likely resulted from a more intense hydrolytic activity, as previously observed in the dark fermentation of sugarcane vinasse (Piffer et al. 2021). The (partial) breakdown of complex structures (such as those found in melanoidins) implies the release of phenolic compounds to the soluble phase, explaining the high participation of PheOH in the residual CODs.
Microbial community composition
The composition of the microbial community samples collected at the beginning (inoculum) and at the end of the operation of the continuous methanogenic reactors (RM1 and RM2 in two different sampling points: FDZ and STB) and further used in MPP tests was determined using 16S rRNA gene amplicon sequencing. Prior to detailing the microbial composition of the samples and making inferences about the primary metabolic pathways, PCoA-based comparisons provided bases to understand differences and similarities among all samples from an overall perspective (Fig. 6), considering the interference of: [i] different regions (FDZ and STB) within a given reactor, [ii] different reactors (RM1 and RM2), [iii] the specialization of the sludge (IN) after long-term (≥ 230 d) operating periods, and [iv] the impacts of sample storage. It is worth highlighting that analyzing the impacts of sample storage was simply a parallel investigation of the study, in an effort to understand potential failures in MPP tests (which were not observed). PCoA results were interpreted considering both the total microbial community (Archaea and Bacteria domains; Fig. 6a) and exclusively the archaeal community (Fig. 6b).
Considering the total microbial community, while little differentiation was observed between different compartments of a given reactor (regardless of the storage in the case of RM2-FDZ), a marked distance was identified between RM1- and RM2-derived samples (Fig. 6a), which were grouped separately. Although the operation of both reactors was finalized under equivalent conditions of OLR (10.0 kg-CODt m−3 d−1), HRT (24.0 h) and alkalinization (NaHCO3 dosing), RM2 was subjected to events of enhanced VFA accumulation when dosing NaOH in previous steps of the operation (Fuess et al. 2021a), which may explain the observed discrepancy. Total microbial communities from both reactors were also very different from IN, considering particularities of each AD system in which samples were collected (IN vs. RM1/RM2): reactor type (UASB vs. AnSTBR), substrate type (vinasse vs. fermented molasses) and OLR (5.0 vs. 10.0 kg-CODt m−3 d−1). In the case of IN, the long-term storage (14 months) impacted the microbial composition to a higher extent than in the case of RM2-FDZ.
Limiting the analysis to the archaeal community (Fig. 6b), samples collected from RM1 and RM2 were very similar, regardless of the compartment. This particular result confirms that variations in the bacterial community of RM2 triggered the discrepancy previously observed in the total microbial communities of both RM1 and RM2 (Fig. 6a). Marked difference was still observed between samples from RM1/RM2 and IN. Interestingly, long-term storage impacted the archaeal community to a much higher extent than the different growth conditions (suspended vs. attached) along the reactors, using the differences between samples RM2-FDZ and RM2-FDZps as the references. Hence, the relative similarity between microbial communities from different compartments of given reactor (regardless of considering only Archaea or both Archaea and Bacteria domains) answers one of the questions raised in Introduction, indicating that a more homogeneous microbial distribution is expected to occur in the biodigestion (methanogenesis) of pre-fermented substrates. However, the unbalanced conversion of organic matter observed in the continuous reactors, i.e. > 70% in the FDZ of both systems (Fuess et al. 2021a), leads to further questions: [i] How metabolically active are the microbial communities located in the bed region? [ii] Could the fixed bed simply be a “cell retention barrier” in the proposed reactor configuration?
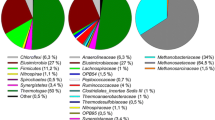
Details of the microbial characterization of the biomass samples at the phylum and genus levels are depicted in Fig. 7, also considering the total microbial community (Archaea and Bacteria domains; Fig. 7a-b) and an isolated analysis of the Archaea domain (Fig. 7c-d). Additionally, heat maps for ASV and genera are presented in the Supplementary data section for further reference. Overall, the Firmicutes phylum prevailed in all samples collected from RM1 and RM2 (Fig. 7a), characterizing a marked different pattern compared to IN and INps. While the phyla Campylobacterota, Firmicutes and Proteobacteria were identified relatively in equivalent abundances in IN, the Proteobacteria phylum prevailed in INps (Fig. 7a). The phylum Euryarchaeota, in which most methanogens are included (Lyu and Liu 2018), was the primary archaeal group identified in all samples (Fig. 7c; except for the case of INps, in which the phylum Halobacterota was also relevant). The Methanothermobacter genus was the most abundant methanogen identified in all samples collected from RM1 and RM2 (relative abundance—RA = 56.7–79.7% among Archaea; Fig. 7d), regardless of the growth condition, i.e. suspended (FDZ) or attached (STB). This result, in association with the identification of the Methanosaeta genus exclusively in samples from RM1 and RM2 (RA = 7.8–25.5% among Archaea; Fig. 7d), largely explain the similarity observed for archaeal communities (pre-storage) from both continuous reactors (Fig. 6b). The storage-dependent difference observed for samples RM2-FDZ and RM2-FDZps resulted from shifts in minor ASV associated with each genus identified. Methanothermobacter also prevailed in the thermophilic sludge (IN) used in the inoculation of RM1 and RM2 (RA = 64.7% among Archaea) and was followed by the Methanoculleus genus as the second most abundant archaeal group (RA = 18.3%) (Fig. 7d). However, the latter (RA = 48.9%) surpassed the first (RA = 41.3%) after the long-term storage of IN (INps; Fig. 7d), which effectively characterized the primary distribution of methanogens during MPP tests. The significant presence of the Methanoculleus genus in both IN and INps samples (and the absence of the Methanosaeta genus) explains their difference relative to RM1 and RM2 (Fig. 6b), whilst the increase in the RA for Methanoculleus in INps explains the distance relative to IN. Methanosarcina (RA < 10.5%) and Bathyarchaeia (RA < 9.0%) genera were also identified in all biomass samples (Fig. 7d), but at lower RA levels compared to the previous groups.
Interestingly, the most abundant methanogens identified in all samples, i.e. Methanothermobacter (Hao et al. 2012; Cheng et al. 2011; Wasserfallen et al. 2000) and Methanoculleus (Dyksma et al. 2020; Manzoor et al. 2016; Shigematsu et al. 2004) genera, grow autotrophically on H2 and CO2, which may sound contradictory when considering two-stage AD schemes. Nearly 90% of the total carbohydrates found in fresh molasses were converted during dark fermentation (Fuess et al. 2021b) prior to feeding RM1 and RM2, markedly eliminating the prompt provision of both H2 and CO2 by acidogenic populations in the methanogenic units. However, the predominance of both genera in the inoculum (combined RA > 80% among Archaea; Fig. 7d) associated with the relatively low availability of acetate (4.6% of the CODs; Table 1) in fermented molasses, i.e. much lower levels than those of lactate (52.9%) and butyrate (17.1%) (Table 1), required the establishment of a strong association between acetogens, syntrophic acetate-oxidizing bacteria (SAOB) and hydrogenotrophic methanogens (HM) in both RM1 and RM2. In particular, the association between SAOB and HM has been systematically pointed to replace acetoclastic methanogenesis under thermophilic conditions (Hao et al. 2012; Cheng et al. 2011; Dyksma et al. 2020), which supports the occurrence of the metabolic sequence proposed. Compared to acetoclastic methanogenesis, methane production from the association between SAOB and HM is thermodynamically favored at high temperatures (Dolfing 2014). In addition, previous investigations indicated the prevalence of the latter pathway at relatively low acetate availability (Shigematsu et al. 2004; Petersen and Ahring 1991), which is in accordance with the metabolite distribution profile in fermented molasses (Table 1).
Differently from the participation of methanogens, notably characterized by a few representative genera (Fig. 7d), a higher diversity of acetogenic and acetate-oxidizing groups at much lower abundance levels was identified. The oxidation of fermentation metabolites into acetate was most likely carried out by the genera Pelotomaculum (RA = 0.9–6.6%, except in IN and INps), Syntrophothermus (RA = 0.6–3.8%, except in IN and INps), Cloacimonadacea W5 (RA = 1.3–5.9%, except in IN and INps), Syntrophomonas (RA = 0.9–4.6%, except in IN and INsp), and Thermodesulfovibrio (RA = 0.3–4.3%, all samples). Butyrate oxidation into acetate was previously associated with the Syntrophothermus (Sekiguchi et al. 2000) and Syntrophomonas (McInerney et al. 1981) genera, whilst both Pelotomaculum (Imachi et al. 2007) and Cloacimonadaceae W5 (Dyksma and Gallert 2019) genera were involved in propionate oxidation. In all cases, syntrophic associations with methanogens were obligate to maintain low acetate levels, characterizing a different aspect of the cases reported in this study, because SAOB most likely consumed most of the available acetate. Interestingly, propionate availability in fermented molasses was much lower than all other fermentation metabolites (0.5% of the CODs; Table 1), so that an intermediate step releasing propionate was most likely established prior to acetogenesis. In particular lactate fermentation into acetate and propionate by species belonging to the Clostridium and Veillonella genera was identified as a relevant pathway in methanogenic environments (Seeliger et al. 2002; Stams et al. 1998), potentially fitting the proposed intermediate step. An ASV associated with the Veillonellales-Senomonadales order was identified primarily in biomass samples collected from the FDZ of both RM1 and RM2 (RA = 6.1–13.5%), which may support this hypothesis. Alternatively, Thermodesulfovibrio may have mediated the syntrophic degradation of lactate, which has been previously reported to occur in both the absence of sulfate and co-culture with hydrogenotrophic methanogens (Sekiguchi et al. 2008). The low sulfate availability in molasses (COD/sulfate > 60.0; Table 1) does not support the long-term persistence of such a sulfate-reducing group in the continuous reactors, suggesting the establishment of an alternative metabolic pathway to maintain cell growth.
Following the SAOB-HM pathway, genera responsible for mediating syntrophic acetate oxidation included Thermoacetogenium (RA = 1.4–2.3%, RM1-FDZ and RM1-STB), Mesotoga (RA = 3.4–7.8%, all samples) and Pseudothermotoga (RA = 0.2–2.1%, except in IN and INps). The Thermoacetogenium genus has been associated with high metabolic flexibility, according to the environmental conditions offered (Hattori et al. 2000). On one hand, acetate oxidation has been reported to occur in co-culture with hydrogenotrophic microorganisms or associated with sulfate reduction in pure culture. On the other hand, acetate production from alcohol fermentation or autotrophically from H2 and CO2 (homoacetogenesis) has also been observed. Acetate oxidation by Pseudothermotoga has been observed only in the presence of thiosulfate or methanogens (Balk et al. 2002), whilst in the case of Mesotoga the acetate-oxidizing pathway is still uncharacterized (Nobu et al. 2015). However, investigations using labelled carbon suggested that the latter syntrophically oxidize acetate only in very low acetate concentrations (Wang et al. 2019).
Concurrently to SAOB, Methanosaeta most likely mediated secondary acetate utilization in methanogenesis via the acetoclastic pathway. Differently from Methanosarcina, which is able to mediate acetoclastic methanogenesis under high acetate availability (Petersen and Ahring 1991; Hori et al. 2006), the activity of Methanosaeta is favored under low acetate concentrations (Petersen and Ahring 1991; Karakashev et al. 2005; Suárez et al. 2018). Hence, efficient consumption of acetate by SAOB (maintained by the high activity of HM) made only a reduced proportion of acetate available for Methanosaeta, characterizing the secondary participation of this group in methane evolution. Interestingly, the highest RA levels observed for Methanosaeta were identified in the STB of both reactors, mainly in the case of RM2 (25.5%; Fig. 7d). Because of VFA accumulation events prior to the finalization of the operation, acetate concentrations higher than 1000 mg-COD L−1 were measured in RM2 (Fuess et al. 2021a), suggesting higher participation of Methanosaeta in methane evolution. From a metabolic perspective, the SAOB-HM pathway was not able to efficiently metabolize excess acetate, requiring the establishment of parallel acetate-consuming pathways. High RA for the Methanosarcina genus was also identified in RM2 (7.4%, FDZ; Fig. 7d), which supports the hypothesis of favoring parallel acetate-consuming pathways. This group may also have participated as a HM, because it includes species capable of growing using any of the known methanogenic pathways, i.e. acetoclastic, hydrogenotrophic, carboxydotrophic (from carbon monoxide), methylotrophic (from methanol, methylamines and methylsulfides) and methyl respiration (methylated compounds + H2) (Buan et al. 2011).
The microbial characterization also provided bases to understand the behavior of methane evolution in MPP tests. The long lag phase periods observed in reactors inoculated with INps (Fig. 2a–c) most likely resulted from non-identifying genera previously pointed out as key-players in acetogenesis (e.g. Pelotomaculum, Syntrophothermus and Cloacimonadaceae W5) and syntrophic acetate oxidation (e.g. Mesotoga and Pseudothermotoga) in the continuous reactors. Null RA values were equally identified for the Methanosaeta genus (Fig. 7d) in INps, suggesting an even stronger dependence of methanogenesis on the SAOB-HM pathway. This hypothesis is corroborated by the delay periods observed in CODs decay when assessing INps (Fig. 3a-c), because an effective participation of acetoclastic methanogenesis would promptly trigger CODs conversion. Because the Thermodesulfovibrio genus was present in INps (at very low initial RA, i.e. 0.3%), this group may have had an important participation in supplying the SAOB-HM pathway on the course of the incubation. Considering RM1- and RM2-derived samples, the minor availability of biomass samples of RM1-FDZ was impeditive to effectively comparing the sludge samples used in MPP tests (post-storage). However, the comparison between RM1-FDZ and RM2-FDZ (both collected during the disassembling of the continuous reactors) indicated higher RA values for key acetogenic (Cloacimonadacea W5, Pelotomaculum, Syntrophothermus and Syntrophomonas), SAOB (Mesotoga, Thermoacetogenium and Pseudothermotoga) and methanogenic (Methanothermobacter) genera in the first sample, explaining the better performances regarding methane evolution when using sludge from RM1. Moreover, the differences in MPP tests most likely did not reach higher levels because the participation of important groups increased in RM2-FDZps (relative to RM2-FDZ), such as the genera Pelotomaculum, Syntrophothermus, Methanothermobacter and Methanosaeta.
In any case, once the association between acetogens, SAOB and HM was efficiently established (regardless of the sludge source), both methane evolution and CODs decay proceeded more efficiently than in previous investigations with both fresh and fermented molasses under mesophilic temperature (30 ºC) (Fuess et al., 2020). In these cases, the dependence of acetoclastic methanogenesis on the activity of acetogens markedly slowed down methane evolution, which occurred at linear (and not exponential) rates in periods longer than 100 h during the incubation (Fuess et al. 2020). Hence, whenever possible, using thermophilic conditions to intentionally select HM may also characterize an essential approach to increase the efficiency of methane production in two-stage AD, because HM present much lower doubling times (a few hours) compared to acetoclastic groups (a few days) (Mosey 1983). The capability of phase separation to increase the robustness of methane evolution relies more on this aspect than on “simply” eliminating enhanced fermentation from the methanogenic unit. Finally, sugar fermentation was most likely carried out by the genera Acetomicrobium (Hania et al. 2016), Anaerolinea (Sekiguchi et al. 2003), Lentimicrobium (Sun et al. 2016), Fervidobacterium (Cai et al. 2007) and Caldicoprobacter (Yokoyama et al. 2010) when using fresh molasses in RM2(Mol) tests, supplying sequential acetogenic and acetate-oxidizing groups.
Conclusions
The primary conclusions drawn from this study include:
-
Phase separation under thermophilic conditions is the best approach to achieve efficient methane production from sugar-rich substrates. In addition to the consolidated idea of minimizing stressful conditions to methanogens by separating enhanced substrate fermentation, the use of high temperatures favor the participation of hydrogenotrophic methanogenesis. In practical aspects, methane evolution will depend on microbial groups (e.g. Methanothermobacter and Methanoculleus genera) that grow faster and are less susceptible to low pH values compared to acetoclastic ones (e.g. Methanosaeta genus). Following the experimental results, the capability of second-stage methanogenic systems to withstand organic loads can be increased by at least threefold compared to single-stage schemes;
-
The microbial community distribution tends to be less stratified in the methanogenic unit of the two-stage biodigestion system, regardless of the differences in both the substrate availability (once the F/M ratio decreases as the liquid flows through the reactor) and the conditions provided for the cell growth (suspended in the bulk liquid or attached to the fixed media). In particular, homogeneity in the distribution of syntrophic acetate-oxidizing bacteria (e.g. Mesotoga and Thermoacetogenium) may be sine qua non for achieving all benefits previously associated with thermophilic two-stage biodigestion (conclusion no. [i]); and,
-
Further studies are still required to better understand the role of microbial communities attached to the fixed bed in such methanogenic reactors. In particular, the scale-up of the AnSTBR may be imperative to unravel the distribution of microbial communities in the different compartments of the system. The concentration of biomass in the feeding zone of bench-scale reactors tends to be proportionally high compared to that of the bed region, triggering a sharp substrate decay which limits defining the effective participation of the attached biomass in the overall substrate conversion.
Abbreviations
- AD:
-
Anaerobic digestion
- AnSTBR:
-
Anaerobic structured-bed reactor
- ASV:
-
Amplicon sequence variant(s)
- CHt:
-
Total carbohydrates
- COD:
-
Chemical oxygen demand
- CODs:
-
Soluble COD
- CODt:
-
Total COD
- F/M:
-
Food-to-microorganism (ratio)
- FDZ:
-
Feeding zone
- fMol:
-
Fermented sugarcane molasses
- GC:
-
Gas chromatography
- HM:
-
Hydrogenotrophic methanogens
- HRT:
-
Hydraulic retention time
- IA:
-
Intermediate alkalinity
- IN:
-
Inoculum (sludge used to inoculate the continuous reactors)
- k1 :
-
First-order kinetic constant
- k1’:
-
Specific first-order kinetic constant
- Mol:
-
Fresh sugarcane molasses
- MPP:
-
Methane production potential
- NCBI:
-
National Center for Biotechnology Information
- OLR:
-
Organic loading rate
- PA:
-
Partial alkalinity
- PCH4 :
-
Potential methane production
- PCoA:
-
Principal coordinates analysis
- PCR:
-
Polymerase chain reaction
- PheOH:
-
Total phenols
- PU:
-
Polyurethane
- PVC:
-
Polyvinyl chloride
- qPCR:
-
Quantitative PCR
- RA:
-
Relative abundance
- RCH4 :
-
Maximum methane production rate
- RCH4’:
-
Specific maximum methane production rate
- RM1/RM2:
-
Nomenclatures for the continuous reactors
- SAOB:
-
Syntrophic acetate-oxidizing bacteria
- SO4 2 − :
-
Sulfate
- sOLR:
-
Specific organic loading rate
- STB:
-
Structured bed
- UASB:
-
Upflow anaerobic sludge blanket (reactor)
- VFA:
-
Volatile fatty acids
- VOAGC :
-
Volatile organic acids by gas chromatography
- VOAtit :
-
Volatile organic acids by titration
- VS:
-
Volatile solids
- λ:
-
Lag phase
References
Adorno MAT, Hirasawa JS, Varesche MBA (2014) Development and validation of two methods to quantify volatile acids (C2–C6) by GC/FID: headspace (automatic and manual) and liquid-liquid extraction (LLE). Am J Anal Chem 5:406–414. https://doi.org/10.4236/ajac.2014.57049
Andersen KS, Kirkegaard RH, Karst SM, Albertsen M (2018) ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 299537. https://doi.org/10.1101/299537
Anzola-Rojas MP, Fonseca SG, Silva CC, Oliveira VM, Zaiat M (2015) The use of the carbon/nitrogen ratio and specific organic loading rate as tools for improving biohydrogen production in fixed-bed reactors. Biotechnol Rep 5:46–54. https://doi.org/10.1016/j.btre.2014.10.010
APHA, Awwa, WEF, (2012) Standard methods for the examination of water and wastewater. APHA, Washington, DC
Aquino SF, Chernicharo CAL, Foresti E, Santos MLF, Monteggia LO (2007) Methodologies for determining the specific methanogenic activity (SMA) in anaerobic sludges. Eng Sanit Ambient 12(2):192–201. https://doi.org/10.1590/S1413-41522007000200010[inPortuguese]
Aquino S, Fuess LT, Pires EC (2017) Media arrangement impacts cell growth in anaerobic fixed-bed reactors treating sugarcane vinasse: structured vs. randomic biomass immobilization. Bioresour Technol 235:219–228. https://doi.org/10.1016/j.biortech.2017.03.120
Balk M, Weijma J, Stams AJM (2002) Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol 52(4):1361–1368. https://doi.org/10.1099/00207713-52-4-1361
Barros VG, Duda RM, Olveira RA (2016) Biomethane production from vinasse in upflow anaerobic sludge blanket reactors inoculated with granular sludge. Braz J Microbiol 47(3):628–639. https://doi.org/10.1016/j.bjm.2016.04.021
Blanco VMC, Fuess LT, Zaiat M (2017) Calcium dosing for the simultaneous control of biomass retention and the enhancement of fermentative biohydrogen production in an innovative fixed-film bioreactor. Int J Hydrogen Energy 42:12181–12196. https://doi.org/10.1016/j.ijhydene.2017.02.180
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Borges AV, Fuess LT, Takeda PY, Alves I, Dias MES, Damianovic MHRZ (2022) Co-digestion of biofuel by-products: Enhanced biofilm formation maintains high organic matter removal when methanogenesis fails. J Environ Manage 310:114768. https://doi.org/10.1016/j.jenvman.2022.114768
Buan N, Kulkarni G, Metcalf W (2011) Genetic methods for Methanosarcina species. Methods Enzymol 494:23–42. https://doi.org/10.1016/B978-0-12-385112-3.00002-0
Buchanan ID, Nicell JA (1997) Model development for horseradish peroxidase catalyzed removal of aqueous phenol. Biotechnol Bioeng 54(3):251–261. https://doi.org/10.1002/(SICI)1097-0290(19970505)54:3%3c251::AID-BIT6%3e3.0.CO;2-E
Cai J, Wang Y, Liu D, Zeng Y, Xue Y, Ma Y, Feng Y (2007) Fervidobacterium changbaicum sp. nov., a novel thermophilic anaerobic bacterium isolated from a hot spring of the Changbai Mountains, China. Int J Syst Evol Microbiol 57(10):2333–2336. https://doi.org/10.1099/ijs.0.64758-0
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Camiloti PR, Mockaitis G, Rodrigues JAD, Damianovic MHRZ, Foresti E, Zaiat M (2014) Innovative anaerobic bioreactor with fixed-structured bed (ABFSB) for simultaneous sulfate reduction and organic matter removal. J Chem Technol Biotechnol 89(7):1044–1050. https://doi.org/10.1002/jctb.4199
Chan YJ, Chong MF, Law CL, Hassell DG (2009) A review on anaerobic-aerobic treatment of industrial and municipal wastewater. Chem Eng J 155(1–2):1–18. https://doi.org/10.1016/j.cej.2009.06.041
Chandra R, Kumar V, Tripathi S (2018) Evaluation of molasses-melanoidin decolourisation by potential bacterial consortium discharged in distillery effluent. 3 Biotech 8:187. https://doi.org/10.1007/s13205-018-1205-3
Cheng L, Dai L, Li X, Zhang H, Lu Y (2011) Isolation and characterization of Methanothermobacter crinale sp. nov., a novel hydrogenotrophic methanogen from the Shengli oil field. Appl Environ Microbiol 77(15):5212–5219. https://doi.org/10.1128/AEM.00210-11
Chernicharo CAL (2007) Anaerobic reactors. IWA Publishing, London. https://doi.org/10.2166/9781780402116
Dolfing J (2014) Thermodynamic constraints on syntrophic acetate oxidation. Appl Environ Microbial 80(4):1539–1541. https://doi.org/10.1128/AEM.03312-13
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric methods for determination of sugar and related substance. Anal Chem 28(3):350–356. https://doi.org/10.1021/ac60111a017
Dyksma S, Gallert C (2019) Candidatus Syntrophosphera thermopropionivorans: a novel player in syntrophic propionate oxidation during anaerobic digestion. Environ Microbiol Rep 11(4):558–570. https://doi.org/10.1111/1758-2229.12759
Dyksma S, Jansen L, Gallert C (2020) Syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of biowaste. Microbiome 8:105. https://doi.org/10.1186/s40168-020-00862-5
Fuess LT, Zaiat M, Nascimento CAO (2020) Molasses vs. juice: Maximizing biohydrogen production in sugarcane biorefineries to diversify renewable energy generation. J Water Process Eng 37:101534. https://doi.org/10.1016/j.jwpe.2020.101534
Fuess LT, Zaiat M, Nascimento CAO (2021a) Thermophilic biodigestion of fermented sugarcane molasses in high-rate structured-bed reactors: Alkalinization strategies define the operating limits. Energy Convers Manage 239:114203. https://doi.org/10.1016/j.enconman.2021.114203
Fuess LT, Fuentes L, Bovio-Winkler P, Eng F, Etchebehere C, Zaiat M, Nascimento CAO (2021b) Full details on continuous biohydrogen production from sugarcane molasses are unraveled: Performance optimization, self-regulation, metabolic correlations and quanti-qualitative biomass characterization. Chem Eng J 414:128934. https://doi.org/10.1016/j.cej.2021.128934
Fuess LT, Cruz RBCM, Zaiat M, Nascimento CAO (2021c) Diversifying the portfolio of sugarcane biorefineries: Anaerobic digestion as the core process for enhanced resource recovery. Renew Sustain Energy Rev 147:111246. https://doi.org/10.1016/j.rser.2021.111246
Hafez H, Nakhla G, El. Naggar MH, Elbeshbishy E, Baghchehsaraee B, (2010) Effect of organic loading on a novel hydrogen bioreactor. Int J Hydrogen Energy 35(1):81–92. https://doi.org/10.1016/j.ijhydene.2009.10.051
Haider MR, Zeshan YS, Malik RN, Visvanathan C (2015) Effect of mixing ratio of food waste and rice husk co-digestion and substrate to inoculum ratio on biogas production. Bioresour Technol 190:451–457. https://doi.org/10.1016/j.biortech.2015.02.105
Hania WB, Bouanane-Darenfed A, Cayol JL, Ollivier B, Fardeau ML (2016) Reclassification of Anaerobaculum mobile, Anaerobaculum thermoterrenum, Anaerobaculum hydrogeniformans as Acetomicrobium mobile comb. nov., Acetomicrobium thermoterrenum comb. nov. and Acetomicrobium hydrogeniformans comb. nov., respectively, and emendation of the genus Acetomicrobium. Int J Syst Evol Microbiol 66(3):1506–1509. https://doi.org/10.1099/ijsem.0.000910
Hao LP, Lü F, Li L, Shao LM, He PJ (2012) Shift of pathways during initiation of thermophilic methanogenesis at different initial pH. Bioresour Technol 126:418–424. https://doi.org/10.1016/j.biortech.2011.12.072
Hattori S, Kamagata Y, Hanada S, Shoun H (2000) Thermoacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol 50(4):1601–1609. https://doi.org/10.1099/00207713-50-4-1601
Holliger C, Alves M, Andrade D, Angelidaki I, Astals S, Baier U et al (2016) Towards a standardization of biomethane potential tests. Water Sci Technol 74(11):2515–2522. https://doi.org/10.2166/wst.2016.336
Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2006) Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl Environ Microbiol 72(2):1623–1630. https://doi.org/10.1128/AEM.72.2.1623-1630.2006
Imachi H, Sakai S, Ohashi A, Harada H, Hanada S, Kagamata Y, Sekiguchi Y (2007) Pelotomaculum propionicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol 57(7):1487–1492. https://doi.org/10.1099/ijs.0.64925-0
Kapp H (1984) Schlammfaulung mit hohem Feststoffgehalt. Stuttgarter Berichte zur Siedlungswasserwirtschaft, Band 86:300, Oldenbourg Verlag, München, Munich, Germany (Sludge with a High Solids Content, Stuttgart reports for Urban Water Management).
Karakashev D, Batstone DJ, Angelidaki I (2005) Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl Environ Microbiol 71(1):331–338. https://doi.org/10.1128/AEM.71.1.331-338.2005
Kiyuna LSM, Fuess LT, Zaiat M (2017) Unraveling the influence of the COD/sulfate ratio on organic matter removal and methane production from the biodigestion of sugarcane vinasse. Bioresour Technol 232:103–112. https://doi.org/10.1016/j.biortech.2017.02.028
Lebrero R, Toledo-Cervantes A, Muñoz R, Del Nery V, Foresti E (2016) Biogas upgrading from vinasse digesters: a comparison between an anoxic biotrickling filter and an algal-bacterial photobioreactor. J Chem Technol Biotechnol 91(9):2488–2495. https://doi.org/10.1002/jctb.4843
Lyu Z, Liu Y (2018) In: Stams AJM, Sousa D (eds) Biogenesis of Hydrocarbons. Handbook of Hydrocarbon and Lipid Microbiology, Springer, Cham, p. 1–59. https://doi.org/10.1007/978-3-319-53114-4_5-1
Manzoor S, Schnürer A, Bongcam-Rudloff E, Müller B (2016) Complete genome sequence of Methanoculleus bourgensis strain MAB1, the syntrophic partner of mesophilic acetate-oxidising bacteria (SAOB). Stand Genomic Sci 11:80. https://doi.org/10.1186/s40793-016-0199-x
McInerney MJ, Bryant MP, Hespell RB, Costerton JW (1981) Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol 41(4):1029–1039. https://doi.org/10.1128/aem.41.4.1029-1039.1981
McMurdie PJ, Holmes S (2013) phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8(4):e61217. https://doi.org/10.1371/journal.pone.0061217
Mockaitis G, Pantoja JLR, Rodrigues JAD, Foresti E, Zaiat M (2014) Continuous anaerobic bioreactor with a fixed-structure bed (ABFSB) for wastewater treatment with low solids and low applied loading content. Bioprocess Biosyst Eng 37:1361–1368. https://doi.org/10.1007/s00449-013-1108-y
Mohana S, Acharya BK, Madamwar D (2009) Distillery spent wash: Treatment technologies and potential applications. J Hazard Mater 163(1):12–25. https://doi.org/10.1016/j.jhazmat.2008.06.079
Moletta R (2005) Winery and distillery wastewater treatment by anaerobic digestion. Water Sci Technol 51(1):137–144. https://doi.org/10.2166/wst.2005.0017
Mosey FE (1983) Mathematical modelling of the anaerobic digestion process: Regulatory mechanisms for the formation of short-chain volatile acids from glucose. Water Sci Technol 15(8–9):209–232. https://doi.org/10.2166/wst.1983.0168
Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu WT (2015) Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9:1710–1722. https://doi.org/10.1038/ismej.2014.256
Oliveira CA, Fuess LT, Soares LA, Damianovic MHRZ (2020) Thermophilic biomethanation of sugarcane molasses comparing single and two-stage systems: Process performance and energetic potential. Bioresour Technol Rep 12:100590. https://doi.org/10.1016/j.biteb.2020.100590
Petersen SP, Ahring BK (1991) Acetate oxidation in a thermophilic anaerobic sewage-sludge digestor: the importance of non-aceticlastic methanogenesis from acetate. FEMS Microbiol Ecol 86(2):149–158. https://doi.org/10.1111/j.1574-6968.1991.tb04804.x
Piffer MA, Zaiat M, Nascimento CAO, Fuess LT (2021) Dynamics of sulfate reduction in the thermophilic dark fermentation of sugarcane vinasse: a biohydrogen-independent approach targeting enhanced bioenergy production. J Environ Chem Eng 9:105956. https://doi.org/10.1016/j.jece.2021.105956
Piffer MA, Oliveira CA, Bovio-Winkler P, Eng F, Etchebehere C, Zaiat M, Nascimento CAO, Fuess LT (2022) Sulfate- and pH driven metabolic flexibility in sugarcane vinasse dark fermentation stimulates biohydrogen evolution, sulfidogenesis or homoacetogenesis. Int J Hydrogen Energy 47(73):31202–31222. https://doi.org/10.1016/j.ijhydene.2022.07.039
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596. https://doi.org/10.1093/nar/gks1219
Ripley LE, Boyle WC, Converse JC (1986) Improved alkalimetric monitoring for anaerobic digestion of high-strength wastes. J Water Pollut Control Fed 58(5):406–411
Rodrigues JAD, Ratusznei SM, Camargo EFM, Zaiat M (2003) Influence of agitation rate on the performance of an anaerobic sequencing batch reactor containing granulated biomass treating low-strength wastewater. Adv Environ Res 7:405–410. https://doi.org/10.1016/S1093-0191(02)00008-4
Santos PS, Zaiat M, Nascimento CAO, Fuess LT (2019) Does sugarcane vinasse composition variability affect the bioenergy yield in anaerobic systems? A dual kinetic-energetic assessment. J Clean Prod 240:118005. https://doi.org/10.1016/j.jclepro.2019.118005
Seeliger S, Janssen PH, Schink B (2002) Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol Lett 211(1):65–70. https://doi.org/10.1111/j.1574-6968.2002.tb11204.x
Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H (2000) Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int J Syst Evol Microbiol 50(2):771–779. https://doi.org/10.1099/00207713-50-2-771
Sekiguchi Y, Yamada T, Hanada S, Ohashi A, Harada H, Kamagata Y (2003) Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int J Syst Evol Microbiol 53(6):1843–1851. https://doi.org/10.1099/ijs.0.02699-0
Sekiguchi Y, Muramatsu M, Imachi H, Narihiro T, Ohashi A, Harada H, Hanada S, Kagamata Y (2008) Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int J Syst Evol Microbiol 58(11):2541–2548. https://doi.org/10.1099/ijs.0.2008/000893-0
Shigematsu T, Tang Y, Kobayashi T, Kawaguchi H, Morimura S, Kida K (2004) Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemist cultivation. Appl Environ Microbiol 70(7):4048–4052. https://doi.org/10.1128/AEM.70.7.4048-4052.2004
Stams AJM, Dijkema C, Plugge CM, Lens P (1998) Contribution of 13C-NMR spectroscopy to the elucidation of pathways of propionate formation and degradation in methanogenic environments. Biodegradation 9(6):463–473. https://doi.org/10.1023/A:1008342130938
Suárez WAB, Vantini JS, Duda RM, Giachetto PF, Cintra LC, Ferro MIT, Oliveira RA (2018) Predominance of syntrophic bacteria, Methanosaeta and Methanoculleus in a two-stage up-flow anaerobic sludge blanket reactor treating coffee processing wastewater at high organic loading rate. Bioresour Technol 268:158–168. https://doi.org/10.1016/j.biortech.2018.06.091
Sun L, Toyonaga M, Ohashi A, Tourlousse DM, Matsuura N, Meng XY, Tamaki H, Hanada S, Cruz R, Yamaguchi T, Sekiguchi Y (2016) Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int J Syst Evol Microbiol 66(7):2635–2642. https://doi.org/10.1099/ijsem.0.001103
Taylor KACC (1996) A simple colorimetric assay for muramic acid and lactic acid. Appl Biochem Biotechnol 56(1):49–58. https://doi.org/10.1007/BF02787869
van Lier JB, van der Zee FP, Frijters CTMJ, Ersahin ME (2015) Celebrating 40 years anaerobic sludge bed reactors for industrial wastewater treatment. Rev Environ Sci Biotechnol 14:681–702. https://doi.org/10.1007/s11157-015-9375-5
Vilela RS, Fuess LT, Saia FT, Silveira CRM, Oliveira CA, Andrade PA, Langenhoff A, van der Zaan B, Cop F, Gregoracci GB, Damianovic MHRZ (2021) Biofuel production from sugarcane molasses in thermophilic anaerobic structured-bed reactors. Renew Sustain Energy Rev 144:110974. https://doi.org/10.1016/j.rser.2021.110974
Wang HZ, Lv XM, Yi Y, Zheng D, Gou M, Nie Y, Hu B, Nobu MK, Narihiro T, Tang YQ (2019) Using DNA-based stable isotope probing to reveal novel propionate- and acetate-oxidizing bacteria in propionate-fed mesophilic anaerobic chemostats. Sci Rep 9:17396. https://doi.org/10.1038/s41598-019-53849-0
Wasserfallen A, Nölling J, Pfister P, Reeve J, Macario EC (2000) Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int J Syst Evol Microbiol 50(1):43–53. https://doi.org/10.1099/00207713-50-1-43
Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York
Yokoyama H, Wagner ID, Wiegel J (2010) Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov. Int J Syst Evol Microbiol 60(1):67–71. https://doi.org/10.1099/ijs.0.011379-0
Zwietering MH, Jongenburger I, Rombouts FM, van’t Riet K (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56(6):1875–1881. https://doi.org/10.1128/aem.56.6.1875-1881.1990
Acknowledgements
This work was supported by the São Paulo Research Foundation (FAPESP) [grant numbers 2017/00080-5, 2015/50684-9, 2015/06246-7 and 2014/50279-4]; and the Coordination for the Improvement of Higher Education Personnel via the Academic Excellence Program (PROEX-CAPES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing interests that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fuess, L.T., Eng, F., Bovio-Winkler, P. et al. Methanogenic consortia from thermophilic molasses-fed structured-bed reactors: microbial characterization and responses to varying food-to-microorganism ratios. Braz. J. Chem. Eng. (2022). https://doi.org/10.1007/s43153-022-00291-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43153-022-00291-x