To the Editor—Although infectivity from respiratory transmission of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) is a well-recognized and evolving area, the role of fecal–oral transmission remains unclear and is a key area requiring further evaluation. SARS-CoV-2 shedding in the gastrointestinal tract (GIT) occurs later than in the respiratory tract. Reference Wu, Guo and Tang1 Here, we report the first case of virus isolation from feces later than respiratory samples and the value of exploring the role of fecal–oral transmission as an underappreciated contributor to infection transmission.
On returning from international travel in March 2020, a 63-year-old man with well-controlled HIV presented to a tertiary-care hospital in Melbourne, Australia, and was subsequently diagnosed with moderate coronavirus disease 2019 (COVID-19). The patient was enrolled through the Sentinel Travelers Research Preparedness Platform for Emerging Infectious Diseases (SETREP-ID, ethics approvals HREC/17/MH/53 and UoM1749349) and provided written informed consent. His CD4 cell count was 973 cells/mm3, and he had had an undetectable HIV viral load (<20 copies/mL) for >5 years. He had a history of ischemic heart disease, stage 3a chronic kidney disease, and previous non-Hodgkin’s lymphoma treated with chemotherapy in 2012.
On admission (4 days after symptom onset), symptoms included diarrhea, history of fever (afebrile on admission), cough with sputum production, chest pain with coughing, myalgia, dyspnea, sore throat, and leg cramps. His oxygen saturation was 99% on room air and his respiratory rate was 30 breaths per minute. The following day his chest pain and diarrhea resolved, but cough and sputum production were ongoing at hospital discharge 5 days later and the following 2 months. He did not receive any specific antiviral or corticosteroid treatment during admission according to the standard of care at the time.
SARS-CoV-2 viral RNA was detected using real-time reverse transcription polymerase chain reaction (RT-PCR) on days 5 and 9 in sputum and on days 6 and 9 in feces. PCR-positive samples were cultured to assess viral infectivity (Supplementary Methods online). Virus was successfully isolated from sputum only on day 5, whereas virus was isolated from feces only on day 9. The cycle threshold (Ct) values for sputum and feces from which virus was isolated were 25 and 23, respectively. Virus isolation was verified from culture medium by a RT-PCR Ct value of 15. By day 53, all samples were RT-PCR negative. At the most recent follow-up (day 116), the patient had made a complete recovery and all samples remained RT-PCR negative (Fig. 1).
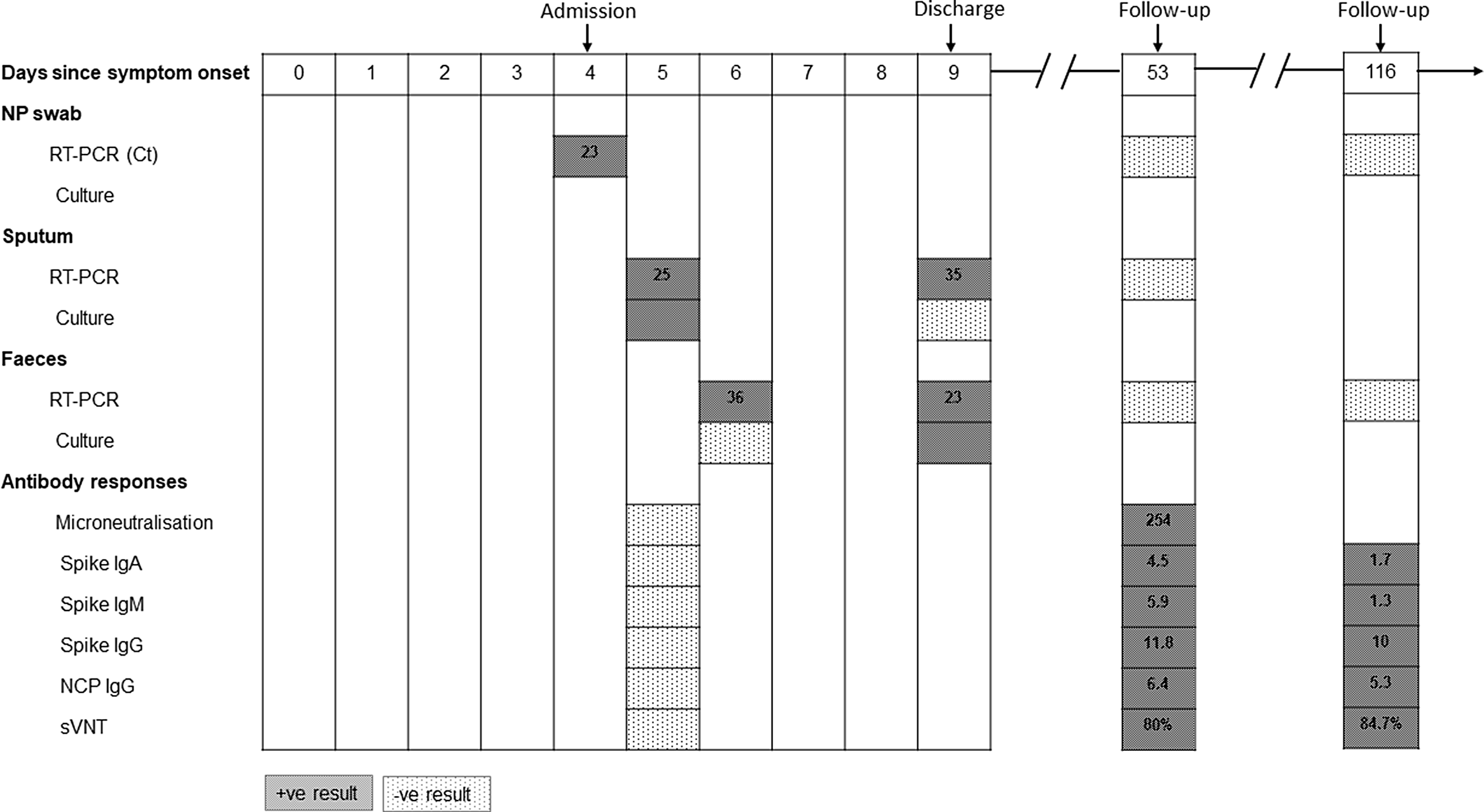
Fig. 1. SARS-CoV-2 shedding, viral isolation, and antibody development in a recovered HIV-positive COVID-19 patient. Red rectangles indicate positive results and green rectangles indicate negative or undetected results. The numbers in RT-PCR rectangles are cycle threshold (Ct) values. Numbers in antibody response rectangles are titers for microneutralization assays and index values for IgA/IgM/IgG. Note. Ig, immunoglobulin; NCP, nucleoscapsid; NP, nasopharyngeal; sVNT, surrogate virus neutralization test.
Neutralizing antibody was measured using 2 methods. The microneutralization assay revealed a rise in titer from undetectable to 254 on paired serum samples (days 5 and 53). The surrogate virus neutralization tests were performed in parallel on samples from days 5, 53, and 116. There was evidence of development of neutralizing antibody by day 53, and persistence of antibodies at day 116 (Fig. 1). IgA, IgM, and IgG responses were positive by day 53 but only IgG persisted at a high level at day 116. In samples from this HIV-positive patient, broad immune activation was observed for both innate and adaptive immune cell subsets at day 5 compared to day 53.
Single-nucleotide polymorphisms (SNPs) were not detected between all samples except the sputum sample from day 9, which had 5 SNPs compared to the other samples. This sample had lower sequence quality (more heterozygous sites/lower genome coverage); thus, whether the genetic differences reflect RNA degradation, sequencing artifacts, or intrahost variation remains unclear.
Understanding the drivers of SARS-CoV-2 transmission is critical to the pandemic response and for implementing key public health measures. The role of fecal–oral transmission is unclear but has been increasingly indicated with the isolation of infectious SARS-CoV-2 in fecal samples. Reference Xiao, Sun and Xu2 Additionally, generation of possible fecal bioaerosols can lead to outbreaks in high-rise buildings Reference Kang, Wei and Yuan3 and hospital settings. Reference McDermott, Alicic, Harden, Cox and Scanlan4 Although studies have shown that SARS-CoV-2 shedding in the GIT occurs later than in the respiratory tract, Reference Wu, Guo and Tang1 this is the first case that demonstrated culturable virus isolation from feces at a later time than respiratory samples.
Although this patient had well-controlled HIV, the link between an altered immune system and viable SARS-CoV-2 in the GIT is unclear. Altered viral transmission dynamics has been observed with influenza and immunosuppression unrelated to HIV infection. Reference Klimov, Rocha, Hayden, Shult, Roumillat and Cox5,Reference Weinstock, Gubareva and Zuccotti6
Importantly, the temporal difference of culturable virus from the respiratory tract and the GIT did not translate to genetic mutations between SARS-CoV-2 virus from the 2 anatomical sites, demonstrating that intrahost variation is unlikely, except for a single lower-quality sequence.
The prevalence of GIT symptoms in SARS-CoV-2 is ˜12%, and viral RNA shedding has been identified in 41% of stool specimens. Reference Parasa, Desai and Thoguluva Chandrasekar7 Angiotensin converting enzyme 2 (ACE-2) is highly expressed in the small intestine, enabling SARS-CoV-2 infection of the digestive system. SARS-CoV-2 has been detected in the intestinal biopsies from asymptomatic individuals 3 months after COVID-19 onset. Reference Gaebler, Wang and Lorenzi8 The focus of infection control and public health measures are on preventing respiratory transmission, and diagnostics used to document viral clearance from the respiratory tract. This case demonstrates that after resolution of diarrhea and most acute symptoms, virus could be cultured from feces. By day 53 of disease onset, feces returned to RT-PCR negative, and although the exact timing is unknown, prolonged respiratory shedding to 60 days after disease onset has been observed. Reference Li, Zhang, Liu and Song9
In summary, we report a case of virus isolation in feces when there was clinical improvement, and viable virus was not detectable in respiratory specimens. Fecal–oral transmission warrants further evaluation, including subgroups such as immunocompromised hosts. Gaining a better understanding of coronavirus disease 2019 (COVID-19) transmission dynamics is critical to this pandemic.
Acknowledgments
We sincerely thank the patient for his ongoing participation in SETREP-ID and for this case report.
Financial support
This work was supported by funding through the National Health and Medical Research Council Centre of Research Excellence (NHMRC CRE), the Australian Partnership for Preparedness Research on Infectious Disease Emergencies (APPRISE AppID 1116530) and in part through funding from the Jack Ma Foundation and the A2 Milk Company. K.S. is supported by an NHMRC Investigator grant. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. L.H. was supported by Melbourne International Research Scholarships and Melbourne International Fee Remission Scholarships from The University of Melbourne.
Conflicts of interest
All authors have no conflicts relevant to this article.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2021.223






