Abstract
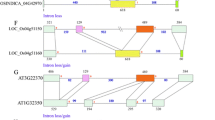
We investigated the copy number of the gene for alternative oxidase (AOX) of Arabidopsis thaliana by amplification by PCR and Southern hybridization. These studies indicated that there are at least four copies of the AOX gene in Arabidopsis. We isolated genomic clones containing individual copies (designated as AOX1a, AOX1b, AOX1c and AOX2) of the AOX genes. Interestingly, two of the AOX genes (AOX1a and AOX1b) were located in tandem in a ca. 5 kb region on one of the chromosomes of Arabidopsis. Comparison between genomic and cDNA sequences of the four AOX genes showed that all AOX genes are divided by three introns and the positions of the introns in AOX1a, AOX1b, AOX1c and AOX2 are the same. We examined whether expression of Arabidopsis AOX genes, like the tobacco AOX1a gene, is enhanced by treatment with antimycin A, an inhibitor of complex III in the mitochondrial respiratory chain. We found that, in young plants, the amount of Arabidopsis AOX1a mRNA was dramatically increased by addition of antimycin A, while the transcription of the other three genes (AOX1b, AOX1c and AOX2) did not respond to antimycin A. Amplification by RT-PCR showed that AOX1a and AOX1c were expressed in all organs examined (flowers and buds, stems, rosette, and roots of 8-week old plants). In contrast, transcripts of AOX1b were detected only in the flowers and buds, and transcripts of AOX2 were detected mainly in stems, rosette and roots. These results suggested that transcriptions of the four genes for alternative oxidase of Arabidopsis are differentially regulated.
Similar content being viewed by others
References
Braun HP, Jänsch L, Kruft V, Schmitz UK: The 'Hinge' protein of cytochrome c reductase from potato lacks the acidic domain and has no cleavable presequence. FEBS Lett 347: 90–94 (1994).
Breathnach R, Chambon P: Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50: 349–383 (1981).
Brown JWS, Smith P, Simpson CG: Arabidopsis consensus intron sequences. Plant Mol Biol 32: 531–535 (1996).
Chaudhuri M, Hill GC: Cloning, sequencing and functional activity of the Trypanosoma brucei brucei alternative oxidase. Mol Biochem Parasitol 83. 125–129 (1996).
Conley CA, Hanson MR: Tissue-specific protein expression in plant mitochondria. Plant Cell 6: 85–91 (1994).
Cruz-Hernández A, Gómez-Lim MA: Alternative oxidase from mango (Mangifera indica L.) is differentially regulated during fruit ripening. Planta 197: 569–576 (1995).
Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT: Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol 22: 497–509 (1995).
de Paepe R, Forchioni A, Chétrit P, Vedel F: Specific mitochondrial proteins in pollen: presence of an additional ATP synthase βsubunit. Proc Natl Acad Sci USA 90: 5934–5938 (1993).
Elthon TE, McIntosh L: Identification of the alternative terminal oxidase of higher plant mitochondria. Proc Natl Acad Sci USA 84: 8399-8403 (1987).
Finnegan PM, Whelan J, Millar AH, Zhang Q, Smith MK, Wiskich JT, Day DA: Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol, in press.
Grohmann L, Rasmusson AG, Heiser V, Thieck O, Brennicke A: The NADH-binding subunit of respiratory chain complex I is nuclear-encoded in plants and identified only in mitochondria. Plant J 10: 793–803 (1996).
Heiser V, Brennicke A, Grohmann L: The plant mitochondrial 22 kDa (PSST) subunit of respiratory chain complex I is encoded by a nuclear gene with enhanced transcript levels in flowers. Plant Mol Biol 31: 1195–1204 (1996).
Hiser C, Kapranov P, McIntosh L: Genetic modification of respiratory capacity in potato. Plant Physiol 110: 277–286 (1996).
Huang J, Struck F, Matzinger DF, Levings III CS: Flowerenhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrial number. Plant Cell 6: 439–448 (1994).
Johns C, Nickels R, McIntosh L, Mackenzie S: The expression of alternative oxidase and alternative respiratory capacity in cytoplasmic male sterile common bean. Sex Plant Reprod 6: 257–265 (1993).
Kearns A, Whelan J, Young S, Elthon TE, Day DA: Tissuespecific expression of the alternative oxidase in soybean and siratro. Plant Physiol 99: 712–717 (1992).
Kingston RE: Guanidinium methods for total RNA preparation. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current Protocols in Molecular Biology, pp. 4.2.1–4.2.8. Greene Publishing Associates/Wiley-Interscience, New York (1991).
Kumar AM, Söll D: Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc Natl Acad Sci USA 89: 10842–10846 (1992).
Lambowitz AM, Sabourin JR, Bertrand H, Nickels R, McIntosh L: Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol Cell Biol 9: 1362–1364 (1989).
Li Q, Ritzel RG, McLean LLT, McIntosh L, Ko T, Bertrand H, Nargang FE: Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics 142: 129–140 (1996).
McIntosh L: Molecular biology of the alternative oxidase. Plant Physiol 105: 781–786 (1994).
Meeuse BJD, Raskin I: Sexual reproduction in arumlily family, with emphasis on thermogenicity. Sex Plant Reprod 1: 3–15 (1988).
Moore AL, Umbach AL, Siedow JN: Structure-function relationships of the alternative oxidase of plant mitochondria: a model of the active site. J Bioenerg Biomembr 27: 367–377 (1995).
Nakagawa T, Maeshima M, Nakamura K, Asahi T: Molecular cloning of a cDNA for the smallest nuclear encoded subunit of sweet potato cytochrome c oxidase. Eur J Biochem 191: 557–561 (1990).
Nordin K, Vahala T, Palva ET: Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21: 641–653 (1993).
Obenland D, Diethelm R, Shibles R, Stewart C: Relationship of alternative respiratory capacity and alternative oxidase amount during soybean seedling growth. Plant Cell Physiol 31: 897–901 (1990).
Rhoads DM, McIntosh L: Isolation and characterization of a cDNA clone encoding an alternative oxidase protein of Sauromatum guttatum (Schott). Proc Natl Acad Sci USA 88: 2122–2126 (1991).
Rhoads DM, McIntosh L: The salicylic acid-inducible alternative oxidase gene aox1 and genes encoding pathogenesis-related proteins share regions of sequence similarity in their promoters. Plant Mol Biol 21: 615–624 (1993).
Sakajo S, Minagawa N, Komiyama T, Yoshimoto A: Molecular cloning of cDNA for antimycin A-inducible mRNA and its role in cyanide-resistant respiration in Hansenula anomala. Biochim Biophys Acta 1090: 102–108 (1991).
Shure M, Wessler S, Fedoroff N: Molecular identification and isolation of the Waxy locus in maize. Cell 35: 225–233 (1983).
Siedow JN, Umbach AL: Plant mitochondrial electron transfer and molecular biology. Plant Cell 7: 821–831 (1995).
Siedow JN, Umbach AL, Moore AL: The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Lett 362: 10–14 (1995).
Stewart CR, Martin BA, Reding L, Cerwick S: Seedling growth, mitochondrial characteristics, and alternative respiratory capacity of corn genotypes differing in cold tolerance. Plant Physiol 92: 761–766 (1990).
Suzuki M, Ario T, Hattori T, Nakamura K, Asahi T: Isolation and characterization of two tightly linked catalase genes from castor bean that are differentially regulated. Plant Mol Biol 25: 507–516 (1994).
Thompson JD, Higgins DG, Gibson TJ: CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673–4680 (1994).
Umbach AL, Siedow JN: Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103: 845–854 (1993).
Umbach AL, Siedow JN: The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that α-keto acid activation involves the formation of a thiohemiacetal. J Biol Chem 271: 25019–25026 (1996).
Vanlerberghe GC, McIntosh L: Lower growth temperature increases alternative pathway capacity and alternative oxidase protein in tobacco. Plant Physiol 100: 115–119 (1992).
Vanlerberghe GC, McIntosh L: Coordinate regulation of cytochrome and alternative pathway respiration in tobacco. Plant Physiol 100: 1846–1851 (1992).
Vanlerberghe GC, McIntosh L: Mitochondrial electron transport regulation of nuclear gene expression: studies with the alternative oxidase gene of tobacco. Plant Physiol 105: 867–874 (1994).
Watanabe N, Nakazono M, Kanno A, Tsutsumi N, Hirai A: Evolutionary variations in DNA sequences transferred from chloroplast genomes to mitochondrial genomes in the Gramineae. Curr Genet 26: 512–518 (1994).
Whelan J, Hugosson M, Glaser E, Day DA: Studies on the import and processing of the alternative oxidase precursor by isolated soybean mitochondria. Plant Mol Biol 27: 769–778 (1995).
Whelan J, McIntosh L, Day DA: Sequencing of a soybean alternative oxidase cDNA clone. Plant Physiol 103: 1481 (1993).
Whelan J, Smith MK, Meijer M, Yu JW, Badger MR, Price GD, Day DA: Cloning of an additional cDNA for alternative oxidase in tobacco. Plant Physiol 107: 1469–1470 (1995).
Whelan J, Millar AH, Day DA: The alternative oxidase is encoded in a multigene family in soybean. Planta 198: 197–201 (1996).
Yamaguchi-Shinozaki K, Shinozaki K: Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236: 331–340 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Saisho, D., Nambara, E., Naito, S. et al. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol 35, 585–596 (1997). https://doi.org/10.1023/A:1005818507743
Issue Date:
DOI: https://doi.org/10.1023/A:1005818507743