Abstract
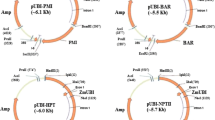
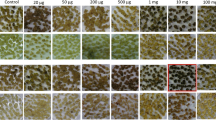
The xylose isomerase gene (xylA) from Thermoanaerobacterium thermosulfurogenes (formerly Clostridium thermosulfurogenes) has been expressed in three plant species (potato, tobacco, and tomato) and transgenic plants have been selected on xylose-containing medium. The xylose isomerase gene was transferred to the target plant by Agrobacterium-mediated transformation. The xylose isomerase gene was expressed using the enhanced cauliflower mosaic virus (CaMV) 35S promoter and the Ω′ translation enhancer sequence from tobacco mosaic virus. Unoptimized selection studies showed that, in potato and tomato, the xylose isomerase selection was more efficient than the established kanamycin resistance selection, whereas in tobacco the opposite was observed. Efficiency may be increased by optimization. The xylose isomerase system enables the transgenic cells to utilize xylose as a carbohydrate source. It is an example of a positive selection system because transgenic cells proliferate while non-transgenic cells are starved but still survive. This contrasts to antibiotic or herbicide resistance where transgenic cells survive on a selective medium but non-transgenic cells are killed. The results give access to a new selection method which is devoid of the disadvantages of antibiotic or herbicide selection.
Similar content being viewed by others
References
ACNFP. Report on the use of antibiotic resistance markers in genetically modified food organisms. Advisory Committee on Novel Foods and Processes, Department of Health and Ministry of Agriculture, Fisheries and Food, London (1994).
Amman E, Brosius J, Ptashne N: Vectors bearing a hydrid trplac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25: 167–178 (1983).
Beck E, Ludwig G, Auerswald, EA, Reiss B, Schaller H: Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19: 327–336 (1982).
Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W: The tomato poly-galacturonase gene and ripening-specific expression in transgenic plants. Plant Mol Biol 11: 651–662 (1988).
Birnboim HC, Doly J: A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucl Acids Res 7: 1513–1523 (1979).
Bojsen K, Donaldson I, Haldrup A, Joersboe M, Kreiberg JD, Nielsen J, Okkels, FT, Petersen SG: Mannose or xylose based positive selection. Patent Application, International Application Number PCT/EP94/00575, International Publication Number WO 94/20627. World Intellectual Property Organization (1994).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of proteindye binding. Anal Biochem 72: 248–254 (1976).
Calgene Inc.: Request for advisory opinion regarding whether the kanamycin resistance gene, a selectable marker, may be used in the production of genetically engineered tomato, cotton, and rapeseed plants intended for human food and animal feed use. FDA Docket Number 90A–0416 (1990).
Callens M, Kersters Hilderson H, Van Opstal O, De Bruyne CK: Catalytic properties of Dxylose isomerase from Streptomyces violaceoruber. Enzyme Microb Technol 8: 696–700 (1986).
David JD, Wiesmeyer H: Control of xylose metabolism in Escherichia coli. Biochim Biophys Acta 201: 497–499 (1970).
Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: Version II. Plant Mol Biol Rep 1: 19–21 (1983).
Feinberg AP, Vogelstein B: A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 (1983).
Flavell, RB, Dart, E, Fuchs, RL, Fraley RT: Selectable marker genes: safe for plants? Bio/technology 10: 141–144 (1992).
Fling ME, Kopf J, Richards C: Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycosidemodifying enzyme, 3″ (9)-O-nucleotidyltransferase. Nucl Acids Res 13: 7095–7106 (1985).
Fuchs RL, Ream JE, Gammond BG, Naylor MW, Leimgruber RM, Berberich SA: Safety assessment of the neomycin phosphotransferase II (NPTII) protein. Bio/technology 11: 1543–1547 (1993).
Hall JC, Kassi PK, Spencer, MS, Born, WHV: An evaluation of the role of ethylene in herbicidal injury induced by picloram or clopyralid in rapeseed and sunflower plants. Plant Physiol 79: 18–23 (1985).
Hodal L, Bochardt A, Nielsen, JE, Mattsson O, Okkels, FT: Detection, expression and specific elimination of endogenous β-glucuronidase activity in transgenic and non-transgenic plants. Plant Sci 87: 115–122 (1992).
Holsters M, DeWaele D, Depicker A, Messens E, Van Montagu M, Schell J: Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163: 181–187 (1978).
Hood EE, Helmer G, Fraley RT, Chilton MD: The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of TDNA. J Bact 168: 1291–1301 (1986).
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT: A simple and general method for transferring genes into plants. Science 227: 1229–1231 (1985).
Itoh Y, Haas D: Cloning vectors derived from the Pseudomonas plasmid pVS1. Gene 36: 2–36 (1985).
Itoh Y, Watson JM, Haas D, Leisinger T: Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid 11: 206–220 (1984).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Joersbo M, Okkels FT: A novel principle for selection of transgenic plant cells: positive selection. Plant Cell Rep 16: 219–221(1996).
Kasai F, Bayer DE: Effects of 2,4-dichlorophenoxyacetic acid, antiauxins, and metabolic perturbations on cytoplasmic and vacuolar pH of corn root tips measured by in vivo 31PNMR. Pestic Biochem Physiol 51: 161–169 (1995).
Kay R, Shan A, Daly M, McPherson J: Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236: 1299–1302 (1987).
Komor E, Thom M, Maretzki: The mechanism of sugar uptake by sugarcane suspension cells. Planta 153: 181–192 (1981).
Lee C, Bagdasarian M, Meng M, Zeikus JG: Catalytic mechanism of xylose (glucose) isomerase from Clostridium thermosulfurogenes. Characterization of the structural gene and function of active site histidine. J Biol Chem 265: 19082–19090 (1990).
Malca I, Endo RM, Long MR: Mechanism of glucose counteraction of inhibition of root elongation by galactose, mannose, and glucosamine. Phytophathology 57: 272–278 (1967).
Murashige T, Skoog F: A revised medium for the rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 (1962).
Nap J-P, Bijvoet J, Stiekema WJ: Biosafety of kanamycin-resistant transgenic plants. Transgen Res 1: 239–249 (1992).
Nissen P, Minocha SC: Inhibition by 2,4-D of somatic embryogenesis in carrot as explored by its reversal by difluoromethyl-ornithine. Physiol Plant 89: 673–680 (1993).
Odell JT, Nagy F, Chua NH: Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promotor. Nature 313: 810–812 (1985).
Okkels FT, Whenham RJ: Method for the selection of genetically transformed cells and compounds for use in the method. International Publication Number WO 93/05163. World Intellectual Property Organization (1992).
Pittard J, Wallace BJ: Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bact 91: 1494–1507 (1966).
Rothstein SJ, Lahners KN, Lotstein RJ, Carozzi NB, Jayne SM, Rice DA: Promoter cassettes, antibiotic-resistance genes, and vectors for plant transformation. Gene 53: 153–161 (1987).
Sambrook J, Fritsch, EF, Maniatis T: Molecular Cloning. A Laboratory Manual, 2nd ed.. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Scharff DA: Biotechnologygene transfer: terminology, techniques, and problems involved. HortScience 26: 1021–1024 (1991).
Shamanna DK, Sanderson KE: Uptake and catabolism of Dxylose in Salmonella typhimurium LTZ. J Bact 139: 64–70 (1979).
Simola LK: The effect of various mono-and disaccharides on the growth of Sphagnum nemoreum thalli in sterile cultures. Physiol Plant 22: 1079–1084 (1969).
Sleat DE, Gallie DR, Jefferson RA, Bevan MW, Turner PC, Wilson TMA: Characterisation of the 5′leader sequence of tobacco mosaic virus RNA as a general enhancer of translation in vitro. Gene 217: 217–225 (1987).
Vancanneyt G, Smidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M: Construction of an introncontaining marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220: 245–250 (1990).
Walden R, Schell J: Techniques in plant molecular biology: progress and problems. Eur J Biochem 192: 563–576 (1990).
Weising K, Schell J, Kahl G: Foreign genes in plants: transfer, structure, expressions, and applications. Annu Rev Genet 22: 421–477 (1988).
WHO: Health aspects of marker genes in genetically modified plants, Report of a WHO workshop, 32 pp. World Health Organization, Food Safety Unit, Geneva (1993).
Yadav NS, Vanderleyden J, Bennett Dr, Barnes WM, Chilton MD: Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci USA 79: 6322–6326 (1982).
Yanisch-Perron C, Vieira J, Messing J: Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 101–119 (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Haldrup, A., Petersen, S.G. & Okkels, F.T. The xylose isomerase gene from Thermoanaerobacterium thermosulfurogenes allows effective selection of transgenic plant cells using D-xylose as the selection agent. Plant Mol Biol 37, 287–296 (1998). https://doi.org/10.1023/A:1005910417789
Issue Date:
DOI: https://doi.org/10.1023/A:1005910417789