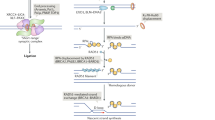
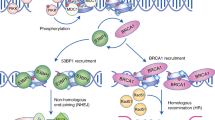
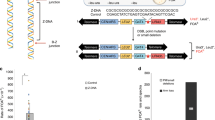
Abstract
Genome stability is of primary importance for the survival and proper functioning of all organisms. Double-stranded breaks in DNA are important threats to genome integrity because they can result in chromosomal aberrations that can affect, simultaneously, many genes, and lead to cell malfunctioning and cell death. These detrimental consequences are counteracted by two mechanistically distinct pathways of double-stranded break repair: homologous recombination and non-homologous end-joining. Recently, unexpected links between these double-stranded break-repair systems, and several human genome instability and cancer predisposition syndromes, have emerged. Now, interactions between both double-stranded break-repair pathways and other cellular processes, such as cell-cycle regulation and replication, are being unveiled.
Key Points
-
DNA double-stranded breaks (DSBs) threaten genome integrity because they cause chromosomal aberrations, leading to loss of cell function and cell death.
-
Two mechanistically distinct, and highly conserved, pathways have evolved to repair DSBs — the homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways. The importance of these pathways in DSB repair is highlighted by the fact that cells with defective HR or NHEJ show widespread chromosomal instability.
-
Recent evidence shows that if endogenously generated DSBs — for example, DSBs created during V(D)J recombination or DNA replication — are not appropriately repaired, they can give rise to chromosomal abnormalities and genomic instability.
-
The proper activation of cell-cycle checkpoints in response to DSBs is also required to prevent chromosomal instability. Chromosomal instability phenotypes can be enhanced by mutations in cell-cycle checkpoint genes.
-
Cellular assays and mouse models are being used to unravel the interactions between components of the DSB-repair pathways and cell-cycle regulators. Work in mice and humans have revealed new links between these repair pathways and certain human cancer predisposition and genome instability syndromes, such as ataxia telangiectasia, Nijmegen breakage syndrome and ataxia telangiectasia-like disorder.
-
Future research is likely to reveal the regulatory networks that coordinate interactions between the different DNA-repair pathways and the cell-cycle checkpoint systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Courtesy of L. R. van Veelen, Department of Cell Biology and Genetics, Erasmus University, Rotterdam, the Netherlands.

Similar content being viewed by others
References
de Boer, J. & Hoeijmakers, J. H. Nucleotide excision repair and human syndromes. Carcinogenesis 21, 453–460 (2000).
Rotman, G. & Shiloh, Y. ATM: from gene to function. Hum. Mol. Genet. 7, 1555–1563 (1998).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instabilities in human cancers. Nature 396, 643–649 ( 1998).
Ramachandran, C. & Melnick, S. J. Multidrug resistance in human tumors — molecular diagnosis and clinical significance. Mol. Diagn. 4, 81–94 ( 1999).
de Klein, A. et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature 300, 765–767 (1982).
Kuzminov, A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16, 373–384 (1995).
Kogoma, T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 (1997).
Dresser, M. E. Meiotic chromosome behavior in Saccharomyces cerevisiae and (mostly) mammals. Mutat. Res. 451, 107– 127 (2000).
Gellert, M. et al. V(D)J recombination: links to transposition and double-strand break repair. Cold Spring Harb. Symp. Quant. Biol. 64, 161–167 (2000).
Papavasiliou, F. N. & Schatz, D. G. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes . Nature 408, 216–221 (2000).
Bross, L. et al. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13, 589– 597 (2000).
Vanasse, G. J., Concannon, P. & Willerford, D. M. Regulated genomic instability and neoplasia in the lymphoid lineage. Blood 94, 3997– 4010 (1999).
Hilgenfeld, E., Padilla-Nash, H., Schrock, E. & Ried, T. Analysis of B-cell neoplasias by spectral karyotyping (SKY). Curr. Top. Microbiol. Immunol. 246, 169– 174 (1999).
Richardson, C. & Jasin, M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405, 697–700 (2000).
Dasika, G. K. et al. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 18 , 7883–7899 (1999).
Zhou, B. B. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000).
Barlow, C. et al. Atm-deficient mice: a paradigm of ataxia telangiectasia . Cell 86, 159–171 (1996).
Elson, A. et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl Acad. Sci. USA 93, 13084 –13089 (1996).
Xu, Y. et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411–2422 (1996).
Liyanage, M. et al. Abnormal rearrangement within the α/δ T-cell receptor locus in lymphomas from Atm-deficient mice. Blood 96, 1940–1946 (2000).
Liao, M. J. & Van Dyke, T. Critical role for Atm in suppressing V(D)J recombination-driven thymic lymphoma. Genes Dev. 13, 1246–1250 (1999).
Petiniot, L. K. et al. Recombinase-activating gene (RAG) 2-mediated V(D)J recombination is not essential for tumorigenesis in Atm-deficient mice. Proc. Natl Acad. Sci. USA 97, 6664– 6669 (2000).References 21 and 22 show that tumorigenesis caused by ATM disruption is delayed when the V(D)J machinery does not make double-stranded breaks in antigen receptor loci. The translocations that can be observed in a RAG-negative background do not involve antigen receptor loci.
Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215– 221 (1992).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858– 5868 (1998).
Paull, T. T. et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 (2000).
Takata, M. et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508 (1998). This paper shows the first example of vertebrate cells with defects in both homologous recombination and non-homologous end-joining. Radiosensitivity and chromosomal aberrations are enhanced when both double-stranded break-repair pathways have been inhibited.
Essers, J. et al. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19, 1703–1710 (2000). This paper shows the synergistic effects of mutations in the genes for DNA-PK CS and Rad54 in mice. Severe combined immunodeficiency (SCID) (DNA-PK CS mutant) mice are hypersensitive to ionizing radiation, whereas Rad54−/− mice are not. However, the double mutant is more sensitive than the SCID single mutant.
Richardson, C. & Jasin, M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 20, 9068–9075 (2000).
Johnson, R. D. & Jasin, M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398– 3407 (2000).
Sung, P., Trujillo, K. M. & Van Komen, S. Recombination factors of Saccharomyces cerevisiae . Mutat. Res. 451, 257– 275 (2000).
Thompson, L. H. & Schild, D. The contribution of homologous recombination in preserving genome integrity in mammalian cells . Biochimie 81, 87–105 (1999).
Kanaar, R., Hoeijmakers, J. H. & van Gent, D. C. Molecular mechanisms of DNA double strand break repair . Trends Cell Biol. 8, 483– 489 (1998).
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349– 404 (1999).
Van Dyck, E., Stasiak, A. Z., Stasiak, A. & West, S. C. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398, 728–731 ( 1999).
Baumann, P. & West, S. C. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23, 247–251 (1998).
Bishop, D. K. et al. Xrcc3 is required for assembly of Rad51 complexes in vivo . J. Biol. Chem. 273, 21482– 21488 (1998).
Takata, M. et al. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20, 6476– 6482 (2000).
Welcsh, P. L., Owens, K. N. & King, M. C. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 16, 69–74 (2000).
Haaf, T., Golub, E. I., Reddy, G., Radding, C. M. & Ward, D. C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes . Proc. Natl Acad. Sci. USA 92, 2298– 2302 (1995).
Tan, T. L. et al. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 9, 325 –328 (1999).
Liu, Y. & Maizels, N. Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO J. 1, 85–90 (2000).
Tashiro, S., Walter, J., Shinohara, A., Kamada, N. & Cremer, T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 150, 283–291 (2000).
Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R. & Bishop, D. K. The breast cancer-susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA crosslinking agent cisplatin. J. Biol. Chem. 275, 23899–23903 ( 2000).
Yuan, S. S. et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 (1999).
Karran, P. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10, 144–150 (2000).
Smith, G. C. & Jackson, S. P. The DNA-dependent protein kinase . Genes Dev. 13, 916–934 (1999).
Barnes, D. E., Stamp, G., Rosewell, I., Denzel, A. & Lindahl, T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8, 1395–1398 (1998).
Frank, K. M. et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396, 173–177 (1998).
Gao, Y. et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95, 891– 902 (1998).
Grawunder, U., Zimmer, D., Fugmann, S., Schwarz, K. & Lieber, M. R. DNA ligase IV is essential for V(D)J recombination and DNA double-stranded break repair in human precursor lymphocytes. Mol. Cell 2, 477–484 ( 1998).
Petrini, J. H. The mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12, 293–296 (2000).
Haber, J. E. The many interfaces of Mre11. Cell 95, 583 –586 (1998).
Morrison, C. & Takeda, S. Genetic analysis of homologous DNA recombination in vertebrate somatic cells. Int. J. Biochem. Cell Biol. 32, 817–831 ( 2000).
Sonoda, E. et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17, 598– 608 (1998).
Tsuzuki, T. et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA 93, 6236–6240 (1996).
Lim, D. & Hasty, P. A mutation in mouse rad51 results in an early embryonic lethal phenotype that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133– 7143 (1996).
Cox, M. M. et al. The importance of repairing stalled replication forks. Nature 404, 37–41 ( 2000).
Flores-Rozas, H. & Kolodner, R. D. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 25, 200–204 (2000).
Kowalczykowski, S. C. Initiation of genetic recombination and recombination-dependent replication . Trends Biochem. Sci. 25, 165– 173 (2000).
Rothstein, R., Michel, B. & Gangloff, S. Replication fork pausing and recombination or 'gimme a break'. Genes Dev. 14, 1– 10 (2000).
Johnson, R. D., Liu, N. & Jasin, M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401, 397– 399 (1999).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Deans, B., Griffin, C. S., Maconochie, M. & Thacker, J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 19, 6675– 6685 (2000).
Griffin, C. S., Simpson, P. J., Wilson, C. R. & Thacker, J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nature Cell Biol. 2, 757– 761 (2000).
Yamaguchi-Iwai, Y. et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18, 6619– 6629 (1999).
Xiao, Y. & Weaver, D. T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25, 2985–2991 ( 1997).
Luo, G. et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA 96, 7376– 7381 (1999).
Morrison, C. et al. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 19, 463– 471 (2000).This report reveals that ATM has an important role in homologous recombination.
Gu, Y., Jin, S., Gao, Y., Weaver, D. T. & Alt, F. W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA 94, 8076–8081 (1997).
Gao, Y. et al. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 9 , 367–376 (1998).
Blunt, T. et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse . Proc. Natl Acad. Sci. USA 93, 10285– 10290 (1996).
Taccioli, G. E. et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity . Immunity 9, 355–366 (1998).
Jhappan, C., Morse, H. C., Fleischmann, R. D., Gottesman, M. M. & Merlino, G. DNA-PKcs : a T-cell tumour suppressor encoded at the mouse scid locus . Nature Genet. 17, 483– 486 (1997).
Nussenzweig, A. et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination . Nature 382, 551–555 (1996).
Zhu, C., Bogue, M. A., Lim, D. S., Hasty, P. & Roth, D. B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86, 379–389 ( 1996).
Vogel, H., Lim, D. S., Karsenty, G., Finegold, M. & Hasty, P. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA 96, 10770 –10775 (1999).
Gu, Y. et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice . Immunity 7, 653–665 (1997).
Ouyang, H. et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 186, 921–929 (1997).
Hsu, H. L. et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14, 2807– 2812 (2000).
Samper, E., Goytisolo, F. A., Slijepcevic, P., van Buul, P. P. W. & Blasco, M. A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO J. 1, 244 –251 (2000).
Frank, K. M. et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5, 993–1002 (2000).
Gao, Y. et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404, 901–904 (2000).
Lee, Y., Barnes, D. E., Lindahl, T. & McKinnon, P. J. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm . Genes Dev. 14, 2576–2580 (2000).References 81 – 83 show that the lethality of mutations in genes for DNA ligase IV or Xrcc4 can be relieved by a mutation in the Trp53 or Atm genes. The double-mutant mice develop T-cell tumours at a very young age, showing the involvement of non-homologus end-joining in tumorigenesis.
Difilippantonio, M. J. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404, 510–514 (2000).
Ferguson, D. O. et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA 97, 6630–6633 (2000).
Karanjawala, Z. E., Grawunder, U., Hsieh, C. L. & Lieber, M. R. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9, 1501 –1504 (1999).References 84 – 86 show the involvement of the Ku heterodimer in the maintenance of chromosome stability, and prevention of tumorigenesis in mice and primary fibroblasts.
Rijkers, T. et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429 (1998).
Essers, J. et al. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89, 195–204 (1997).
Tutt, A. et al. Absence of BRCA2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9, 1107–1110 ( 1999).
Yu, V. P. et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14, 1400–1406 ( 2000).
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999).
Snouwaert, J. N. et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene 18, 7900–7907 ( 1999).References 91 and 92 report the involvement of the Brca1 gene in homologous recombination.
Friedman, L. S. et al. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 58, 1338– 1343 (1998).
Connor, F. et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nature Genet. 17, 423 –430 (1997).
Riballo, E. et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 9, 699– 702 (1999).
Jeggo, P. A., Carr, A. M. & Lehmann, A. R. Splitting the ATM: distinct repair and checkpoint defects in ataxia-telangiectasia. Trends Genet. 14, 312–316 (1998).
Zhao, S. et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405, 473– 477 (2000).
Wu, X. et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405, 477 –482 (2000).
Lim, D. S. et al. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway . Nature 404, 613–617 (2000).
Gatei, M. et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nature Genet. 25, 115– 119 (2000).References 97 – 100 , which report the ATM-dependent phosphorylation of the NBS1 protein under ionizing radiation, explain the similar phenotypes of ATM- and NBS1-deficient cells.
Stewart, G. S. et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587 ( 1999).
Karow, J. K., Wu, L. & Hickson, I. D. RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev. 10, 32–38 (2000).
Wang, W. et al. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 19, 3428 –3435 (2000).
Cooper, M. P. et al. Ku complex interacts with and stimulates the Werner protein . Genes Dev. 14, 907–912 (2000).
Li, B. & Comai, L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem. 275, 28349–28352 ( 2000).
Cortez, D., Wang, Y., Qin, J. & Elledge, S. J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286, 1162– 1166 (1999).
Banks, R. E. et al. Proteomics: new perspectives, new biomedical opportunities . Lancet 356, 1749–1756 (2000).
Gygi, S. P. & Aebersold, R. Mass spectrometry and proteomics . Curr. Opin. Chem. Biol. 4, 489– 494 (2000).
Tibbetts, R. S. et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14, 2989–3002 (2000).
Fulop, G. M. & Phillips, R. A. The scid mutation in mice causes a general defect in DNA repair. Nature 347, 479–482 (1990).
Nacht, M. et al. Mutations in the p53 and SCID genes cooperate in tumorigenesis . Genes Dev. 10, 2055–2066 (1996).
Li, G. C. et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2, 1– 8 (1998).
Yamaguchi-Iwai, Y. et al. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18, 6430–6435 (1998).
Ludwig, T., Chapman, D. L., Papaioannou, V. E. & Efstratiadis, A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11, 1226– 1241 (1997).
Sharan, S. K. et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810 (1997).
Acknowledgements
The authors thank members of the Department of Cell Biology and Genetics for many useful comments on the manuscript. D.C.v.G. is a fellow of the Royal Netherlands Academy of Arts and Sciences (KNAW). Research in our department is supported by the Dutch Cancer Society, the Netherlands Organization for Scientific Research and the European Commission.
Author information
Authors and Affiliations
Related links
Related links
DATABASE LINKS
ataxia telangiectasia-like disorder
FURTHER INFORMATION
Tokyo Medical University's animations of chromosomal structural abnormalities
Glossary
- NUCLEOTIDE EXCISION REPAIR
-
(NER). A DNA-repair pathway that removes ultraviolet-light-induced DNA damage (such as thymidine dimers) and bulky DNA adducts by excising the oligonucleotide that contains the damaged base(s). The single-stranded gap is filled in by using the intact strand as a template.
- MISMATCH REPAIR
-
(MMR). A DNA-repair pathway that removes mismatched bases and corrects the insertion or deletion of short stretches of (repeated) DNA.
- PHILADELPHIA CHROMOSOME
-
Chromosome 22, from which the tip of the q arm has been exchanged for a small region of the q arm of chromosome 9, fusing together the BCR and ABL1 genes.
- SPECTRAL KARYOTYPING
-
(SKY). A karyotyping method that allows all the chromosomes of an organism to be identified in a single metaphase spread. Each chromosome is labelled with chromosome-specific probes that can be visualized as different colours. This technique is useful for identifying certain chromosomal abnormalities.
- IMMUNOGLOBULIN
-
(Ig). Antigen-receptor molecules produced by B cells that consist of two heavy chains and two light chains.
- T-CELL RECEPTOR
-
(Tcr). Antigen-receptor molecules produced by T cells that consist of either α and β, or γ and δ chains.
- METAPHASE SPREAD
-
A collection of chromosomes arrested at the prophase of mitosis. Because the chromosomes are highly condensed at this stage of cell division, they are visible under a light microscope.
- ISOTYPE
-
Class of immunoglobulin (Ig) protein that is determined by the constant region of the IgH gene that is placed nearest to the joining (J) segments. The isotype can switch during development of the B cell.
- PARALOGUE
-
A locus that is homologous to another in the same genome.
- RAD51 NUCLEOPROTEIN FILAMENT
-
A helical filament of RAD51 protein that covers single-stranded DNA. It contains approximately three bases for every RAD51 monomer and six monomers per helical turn. The nucleoprotein filament can pair with homologous double-stranded DNA.
- MACROCHROMOSOME
-
The chicken genome is divided into macro- and minichromosomes. Only macrochromosomes are large enough to be easily observable under a light microscope.
- LATENCY TIME
-
The time required to develop visible signs of disease, for example, a tumour.
- HYPOMORPHIC MUTATION
-
A mutation that does not completely eliminate the wild-type function of a gene and gives a less severe phenotype than a loss-of-function mutation.
Rights and permissions
About this article
Cite this article
van Gent, D., Hoeijmakers, J. & Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2, 196–206 (2001). https://doi.org/10.1038/35056049
Issue Date:
DOI: https://doi.org/10.1038/35056049
This article is cited by
-
Angle modulated two-dimensional single cell pulsed-field gel electrophoresis for detecting early symptoms of DNA fragmentation in human sperm nuclei
Scientific Reports (2024)
-
Long-term outcomes of young, node-negative, chemotherapy-naïve, triple-negative breast cancer patients according to BRCA1 status
BMC Medicine (2024)
-
Radiation makes cells select the form of death dependent on external or internal exposure: apoptosis or pyroptosis
Scientific Reports (2023)
-
TYMS promotes genomic instability and tumor progression in Ink4a/Arf null background
Oncogene (2023)
-
Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions
Signal Transduction and Targeted Therapy (2023)