Abstract
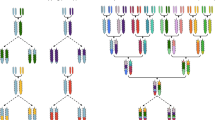
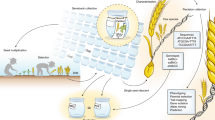
Naturally occurring variation among wild relatives of cultivated crops is an under-exploited resource in plant breeding. Here, I argue that exotic libraries, which consist of marker-defined genomic regions taken from wild species and introgressed onto the background of elite crop lines, provide plant breeders with an important opportunity to improve the agricultural performance of modern crop varieties. These libraries can also act as reagents for the discovery and characterization of genes that underlie traits of agricultural value.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Khush, G. S. Green Revolution: the way forward. Nature Rev. Genet. 2, 815–822 (2001).
Peng, J. et al. 'Green Revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999).
Lev-Yadun, S., Gopher, A. & Abbo, S. Archaeology. The cradle of agriculture. Science 288, 1602–1603 (2000).
Ladizinsky, G. Plant Evolution under Domestication (Kluwer Academic, Dordrecht, The Netherlands, 1998).
Jarret, R. L. & Newman, M. Phylogenetic relationships among species of Citrullus and the placement of C. rehmii De Winter as determined by internal transcribed spacer (ITS) sequence heterogeneity. Genet. Res. Crop Evol. 47, 215–222 (2000).
Koornneef, M. & Stam, P. Changing paradigms in plant breeding. Plant Physiol. 125, 156–159 (2001).
Bessey, C. E. Crop improvement by utilizing wild species. Am. Breed. Assoc. II, 112–118 (1906).
Fedak, G. Molecular aids for integration of alien chromatin through wide crosses. Genome 42, 584–591 (1999).
Villareal, R. L., Del Toro, E., Mujeeb-Kazi, A. & Rajaram, S. The 1BL/1RS chromosome translocation effect on yield characteristics in a Triticum aestivum L. cross. Plant Breed. 114, 497–500 (1995).
Hoisington, D. et al. Plant genetic resources: what can they contribute toward increased crop productivity? Proc. Natl Acad. Sci. USA 96, 5937–5943 (1999).
Kovacs, M. I. P., Howes, N. K., Clarke, J. M. & Leisle, D. Quality characteristics of durum wheat lines deriving high protein from Triticum dicoccoides (6b) substitution. J. Cereal Sci. 27, 47–51 (1998).
Pan, Q. et al. Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155, 309–322 (2000).
Rick, C. M. High soluble-solids content in large-fruited tomato lines derived from a wild green-fruited species. Hilgardia 42, 493–510 (1974).
Ronen, G., Carmel-Goren, L., Zamir, D. & Hirschberg, J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of β- and old-gold color mutations in tomato. Proc. Natl Acad. Sci. USA 97, 11102–11107 (2000).
Plunknett, D. L., Smith, N. J. H., Williams, J. T. & Murthi-Anishetty, N. Gene Banks and the World's Food (Princeton Univ. Press, Princeton, New Jersey, 1987).
Brar, D. S. & Khush, G. S. Alien introgression in rice. Plant Mol. Biol. 35, 35–47 (1997).
Moncada, P. et al. Quantitative trait loci for yield and yield components in an Oryza sativa x O. rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102, 41–52 (2001).
Yano, M. Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4, 130–135 (2001).
Singh, R. J. & Hymowitz, T. Soybean genetic resources and crop improvement. Genome 42, 605–616 (1999).
Sebolt, A. M., Shoemaker, R. C. & Diers, B. W. Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci. 40, 1438–1444 (2000).
Small, R. L., Ryburn, J. A. & Wendel, J. F. Low levels of nucleotide diversity at homoeologous Adh loci in allotetraploid cotton (Gossypium L.). Mol. Biol. Evol. 16, 491–501 (1999).
Iqbal, M. J., Reddy, O. U. K., El-Zik, K. M. & Pepper, A. E. A genetic bottleneck in the 'evolution under domestication' of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor. Appl. Genet. 103, 547–554 (2001).
McCarty, J. C. & Percy, R. G. in Genetic Improvement of Cotton: Emerging Technologies (eds Jenkins, J. N. & Saha, S.) 65–79 (USDA — Agricultural Research Service: Science, Enfield, New Hampshire, 2001).
Doebley, J. & Stec, A. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134, 559–570 (1993).
Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. The limits of selection during maize domestication. Nature 398, 236–239 (1999).
Mauricio, R. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nature Rev. Genet. 2, 370–381 (2001).
Ray, J. D., Kindiger, B. & Sinclair, T. R. Introgressing root aerenchyma into maize. Maydica 44, 113–117 (1999).
Tanksley, S. D. & McCouch, S. R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066 (1997).
Lee, M. Genome projects and gene pools: new germplasm for plant breeding? Proc. Natl Acad. Sci. USA 95, 2001–2004 (1998).
Fulton, T. M. et al. Advanced backcross QTL analysis of a Lycopersicon esculentum x L. parviflorum cross. Theor. Appl. Genet. 100, 1025–1042 (2000).
Eshed, Y. & Zamir, D. Less than additive epistatic interactions of QTL in tomato. Genetics 143, 1807–1817 (1996).
Kuspira, J. & Unrau, J. Genetic analysis of certain characters in common wheat using all chromosome substitution lines. Can. J. Plant Sci. 37, 300–326 (1957).
Eshed, Y. & Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995).
Fridman, E., Pleban, T. & Zamir, D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl Acad. Sci. USA 97, 4718–4723 (2000).
Ramsay, L. D. et al. The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39, 558–567 (1996).
Young, N. D. A cautiously optimistic vision for marker-assisted breeding. Mol. Breed. 5, 505–510 (1999).
Roessner, U. et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001).
Gale, M. D. & Devos, K. M. Plant comparative genetics after 10 years. Science 282, 656–659 (1998).
Patterson, A. H. et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269, 1714–1717 (1995).
Zhong, G. Y. Genetic issues and pitfalls in transgenic plant breeding. Euphytica 118, 137–144 (2001).
Uzogara, S. G. The impact of genetic modification of human foods in the 21st century: a review. Biotechnol. Adv. 18, 179–206 (2000).
Mann, C. C. Crop scientists seek a new revolution. Science 283, 310–314 (1999).
Acdemy, N. Transgenic Plants and World Agriculture (National Academy Press, Washington DC, 2000).
Mitten, D. H., MacDonald, R. & Klonus, D. Regulation of foods derived from genetically engineered crops. Curr. Opin. Biotechnol. 10, 298–302 (1999).
Kuiper, H. A., Kleter, G. A., Noteborn, H. P. J. M. & Kok, E. J. Assessment of food safety issues related to genetically modified foods. Plant J. 27, 503–528 (2001).
Acknowledgements
I thank E. Fridman, Y. Eshed and S. Abbo for helpful discussions. This work was supported by the United States–Israel Binational Research and Development Fund (BARD).
Author information
Authors and Affiliations
Related links
Glossary
- ACCESSION
-
A sample of plant material that is collected at a specific location and maintained in a seed bank.
- ELITE VARIETY
-
A variety that excels under conditions of modern intensive agriculture.
- EPISTASIS
-
An interaction between non-allelic genes, such that one gene masks, interferes with or enhances the expression of the other gene.
- HETEROSIS
-
Hybrid vigour that leads to superior crop varieties.
- INTROGRESSION BREEDING
-
The incorporation of selected traits from an unadapted exotic resource through a succession of crosses (backcrosses) to a commercially elite variety.
- LANDRACE
-
A locally adapted, cultivated variety that is selected by farmers.
- LODGING
-
The collapse of top-heavy plants, particularly grain crops.
- PYRAMIDING
-
The accumulation of several independent traits in the same genotype through introgression breeding.
- QUANTITATIVE TRAIT LOCI
-
(QTL). Genetic loci that are identified through the statistical analysis of complex traits (such as plant height or body weight). Quantitative traits are typically affected by more than one gene and by the environment.
Rights and permissions
About this article
Cite this article
Zamir, D. Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2, 983–989 (2001). https://doi.org/10.1038/35103590
Issue Date:
DOI: https://doi.org/10.1038/35103590
This article is cited by
-
Transcriptomes of developing fruit of cultivated and wild tomato species
Molecular Horticulture (2023)
-
Tomato super-pangenome highlights the potential use of wild relatives in tomato breeding
Nature Genetics (2023)
-
Genetic diversity and conservation of Siberian apricot (Prunus sibirica L.) based on microsatellite markers
Scientific Reports (2023)
-
Genetic mapping and prediction for novel lesion mimic in maize demonstrates quantitative effects from genetic background, environment and epistasis
Theoretical and Applied Genetics (2023)
-
A callus-derived regeneration and Agrobacterium-mediated gene transformation developed for bilberry, Vaccinium myrtillus
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)