Abstract
Duplications in the ~2 Mb desert region upstream of SOX9 at 17q24.3 may result in familial 46,XX disorders of sex development (DSD) without any effects on the XY background. A balanced translocation with its breakpoint falling within the same region has also been described in one XX DSD subject. We analyzed, by conventional and molecular cytogenetics, 19 novel SRY-negative unrelated 46,XX subjects both familial and sporadic, with isolated DSD. One of them had a de novo reciprocal t(11;17) translocation. Two cases carried partially overlapping 17q24.3 duplications ~500 kb upstream of SOX9, both inherited from their normal fathers. Breakpoints cloning showed that both duplications were in tandem, whereas the 17q in the reciprocal translocation was broken at ~800 kb upstream of SOX9, which is not only close to a previously described 46,XX DSD translocation, but also to translocations without any effects on the gonadal development. A further XX male, ascertained because of intellectual disability, carried a de novo cryptic duplication at Xq27.1, involving SOX3. CNVs involving SOX3 or its flanking regions have been reported in four XX DSD subjects. Collectively in our cohort of 19 novel cases of SRY-negative 46,XX DSD, the duplications upstream of SOX9 account for ~10.5% of the cases, and are responsible for the disease phenotype, even when inherited from a normal father. Translocations interrupting this region may also affect the gonadal development, possibly depending on the chromatin context of the recipient chromosome. SOX3 duplications may substitute SRY in some XX subjects.
Similar content being viewed by others
Introduction
46,XX disorders of sex development (DSDs) are congenital conditions in which, in the presence of a female karyotype, the development of gonadal and anatomical sex is atypical, ranging from various degrees of ambiguous genitalia to phenotypic males with azoospermia. These conditions are poorly characterized, at least in subjects whose DNA does not contain SRY, the gene triggering testis differentiation in mammals.1 In fact, in most XX males, SRY is transposed to the tip of Xp as a consequence of a recurrent Xp;Yp translocation, arising predominantly by nonallelic homologous recombination between PRKX and PRKY on a particular Y haplotypic background.2, 3 These males, usually with small testes, are essentially picked up among men with nonobstructive azoospermia.
A much less well-understood category is that of the 46,XX DSDs negative for SRY. Recently, six of these cases have been reported carrying partially overlapping amplifications of a gene-desert region located ~500 kb upstream of SOX9.4, 5, 6, 7 It has been proposed that these CNVs could be responsible for altered expression of SOX9 in the developing gonad on an XX background. Some of the reported cases represent familial 46,XX DSDs, as normal and fully fertile XY fathers can carry this CNV.
In the XY early gonad, after initial activation by SRY, SOX9 maintains its expression thanks to a positive feedback loop triggering the pathway of testis differentiation.8 Actually, SOX9 transgenic expression in XX gonads is sufficient to induce testis formation in mice.9 Moreover, a single case of an XX boy is reported with a >11 Mb duplication, including SOX9.10 SOX9 is also involved in several processes during the embryo development, and defects of this gene are responsible for campomelic dysplasia with/without XY sex reversal (OMIM:114290). This gene has a large upstream desert region of ~2 Mb enriched in tissue- and time-specific regulatory elements, as it can be deduced by different abnormal phenotypes associated with CNVs or reciprocal translocations, interrupting the region itself. Among them, isolated Pierre Robin sequence (PRS) (OMIM:261800), congenital heart defects, and XY sex reversal have been associated mainly with deletions of the region, and in few cases with reciprocal translocations,4, 11, 12, 13 whereas duplications have been reported both in familial cases of brachydactyly-anonychia14 and in the abovementioned XX subjects with DSD. In the latter category, a single case with a reciprocal translocation, interrupting the desert region ~800 kb upstream of SOX9, has also been described.15
Recently, a role in the developing gonad has also been proposed for SOX3 (Sry-related HMG box-containing gene 3), a gene closely related to both SOX9 and SRY. Ectopic expression of Sox3 in the mouse bipotential gonad frequently leads to complete XX sex reversal.16 Moreover, duplications of SOX3 or its 5′ region have been reported in 46,XX DSD patients.16, 17
Here, we present two new subjects with cryptic duplications upstream of SOX9 that were identified among 19 unrelated novel cases with 46,XX isolated DSDs, either familial (2 cases) or sporadic (17 cases), all SRY-negative. In the same cohort, we also identified the second case of reciprocal translocation upstream of SOX9, causing XX DSD. A further 46,XX boy with intellectual disability (ID) and a cryptic duplication, including SOX3, is also presented.
Our data confirm that CNVs and structural rearrangements (cases 1–3) involving desert regions are responsible for the developmental defects by dysregulation of non-coding cis-regulatory elements, 18 and that 46,XX SRY-negative DSD individuals present a duplication upstream of SOX9 in at least 10.5% of cases. The role of CNVs in gonadal disorders is further stressed by case 20.
Subjects and methods
Patient samples
Patients were collected during >20 years. The study was approved by the Ethical Committee of the University of Pavia. Written informed consent was obtained from all patients.
Conventional and molecular cytogenetics
Conventional cytogenetics was done on GTG-banded metaphases. Molecular karyotyping was performed for cases 1–3 and for all cases reported in Supplementary Table 1 by using oligonucleotide array-CGH platforms (180 K SurePrint G3 Human Kit, Agilent Technologies, Santa Clara, CA, USA). For case 20, the trio analysis was performed using the Genome Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). For details, see Supplementary Material. FISH experiments were performed on case 3, as reported, 19 by using the following probes: RP11-238F2, RP11-589A10, CTD-2652P12, RP11-879D6, RP11-474K15, RP11-676K3, RP11-13H17, RP11-661B15, RP11-103P20, RP11-36H11. SRY analysis was done as reported in Supplementary Material.
The observed CNVs have been submitted to the public database DECIPHER (http://decipher.sanger.ac.uk; IDs from case 1–case 20: 293610, 293615, 293631, 293646).
X-inactivation analysis
X-inactivation analysis was performed for case 20 according to Allen et al.20
Breakpoint mapping
For cases 1 and 2, we used quantitative PCR (qPCR) to verify and restrict the breakpoint regions characterized by array-CGH, followed by long-range PCR.21 To clone case 3 breakpoint junctions, we used pooled long-range PCR reactions (for details, see Supplementary Material). Target sequences for qPCR analysis were selected using the Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA). Long-range PCRs were performed with JumpStart Red ACCUTaq LA DNA polymerase (Sigma-Aldrich, St Louis, MO, USA). PCR products were analyzed on 0.8% agarose TAE gels. The UCSC Genome Browser (hg19 assembly) maps and sequence were used as reference. Sequencing reactions were performed with a Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and run on an ABI Prism 3500DX/XL Genetic Analyzer.
In silico TFBSs evaluation
For case 3, the presence of potential transcription factor-binding sites (TFBSs) altered by the translocation breakpoints was evaluated by using the TFSEARCH tool (http://www.rwcp.or.jp/papia/).22 In cases 1–3, the region involved by the rearrangement was also evaluated in the light of integrated regulation from ENCODE, HMR-conserved TFBSs and human body map lincRNAs, and TUCP transcripts tracks embedded in the UCSC genome browser (http://genome.ucsc.edu/).
Results
Clinical description
Case 1
The patient, a normal adult male, was investigated because of infertility. Physical examination showed normal male secondary sexual characteristics and bilateral gynecomastia. Laboratory investigations showed azoospermia, low serum testosterone, and increased FSH and LH.
Case 2
The patient was born at term after two normal female children from nonconsanguineous parents. A threatened abortion in the first trimester is documented. External genitalia were ambiguous with hypertrophic clitoris, single meatus, and urogenital sinus. Ecography showed absent uterus, vaginal atresia, and two ovoidal gonads detected in the inguinal canal. At 8 months of age, hormonal values were as the following: LH was below 0.1 mU/ml (normal range <0.1–6 mU/ml), FSH was 0.8 mU/ml (normal range <0.1–18 mU/ml), testosterone was below 10 ng/dl (normal ranges: 12–21 ng/dl males, 6–82 ng/dl females), androstenedione was below 30 ng/dl (normal range 40–260 ng/dl), estradiol was below 25 pg/ml (normal values <25 pg/ml), anti-Müllerian hormone was above 21 ng/ml (normal ranges: 84–141 ng/ml males,<0.14 ng/ml females). The testosterone response to human chorionic gonadotropin (hHCG) administration was low: 76 ng/dl (normal values >100 ng/dl). After hHCG stimulation, the left gonad showed the presence of follicles and assumed the aspect of an ovotestis at echography, whereas the right one appeared as a testicle. Bilateral gonadal biopsy was performed at 1 year of age. Both gonads were presented with the caudal portions macroscopically compatible with male gonads and the cranial ones with female gonads. At histological examination, the caudal segments showed testicular tissue with prepubertal seminiferous tubules, whereas the cranial ones showed ovarian tissue with numerous oocytes (Supplementary Figure 1).
Case 3
The patient was ascertained in adult age because of infertility. His height was 171 cm, between the 10th and the 25th centile, much shorter than expected on the basis of the mid-parental height (183 cm). He presented with infertility and erectile dysfunction, loss of libido, and asthenia. On general examination, the patient had mild dysmorphisms, such as micrognathia, hypertelorism, short neck. He had bilateral testicular hypotrophy, normal penis, and absence of gynecomastia. FSH (53.3 mU/ml, normal values 0.7–11.1) as well as LH (19.4 mU/ml, normal values 0.8–8.0) and androstenedione (13.1 nmol/l, normal values 2.1–10.8) levels were elevated, whereas he had low serum testosterone (210 ng/dl, normal values 260–1600). The patient also suffered from osteopenia.
Cases 4–19
These cases, all with normal 46,XX karyotype and SRY-negative, were ascertained because of ambiguous genitalia, or hypogonadism, or azoospermia. In all of them, genomic arrays did not show any significant CNV. Their clinical details are summarized in Supplementary Table 1. Some of them, previously published, have now been tested by array-CGH.
Case 20
The patient was ascertained at 8 years of age because of mild developmental and language delay. He was born at term from nonconsanguineous parents, after an uncomplicated pregnancy. At birth, he weighed 3240 g (75th centile) and his length and head circumference were 49 cm (50th centile) and 35 cm (50–75th centile), respectively. He started walking and talking after 2 years of age. No facial or skeletal abnormalities were identified by physical examination, and he had normal male genitalia. The parents reported sleep disturbances with some episodes of pavor nocturnus.
Molecular cytogenetic investigations
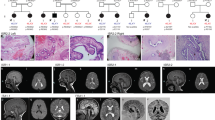
For all the reported cases, except for case 3, a normal 46,XX karyotype was identified by conventional cytogenetics. The presence of SRY was ruled out in all patients by PCR or FISH analysis. Genome-wide copy number analysis was performed by CGH- or SNP-arrays for all reported patients. The results for cases 1–3 and 20 together with their clinical data are summarized in Table 1. In cases 1 and 2, array-CGH analysis identified partially overlapping 17q24.3 duplications of different sizes, involving the gene-desert region upstream of SOX9 (Figure 1a). For both cases, the duplication was also present in paternal DNA. Both sisters of case 2 did not show the RevSex duplication.
Genomic rearrangements in cases 1, 2, and 20. (a) Duplications of partially overlapping 17q24.3 regions in cases 1 (upper panel) and 2 (lower panel): an enlargement of a 2.05-Mb region from chromosome 17 profile is shown, with the duplications highlighted by the shaded areas; (b) DNA sequences spanning the chromosome 17 duplication breakpoints in both cases aligned with the reference sequences; (c) SNP-array profile of chromosome X from case 20 showing the 5.6-Mb duplication encompassing SOX3; (d) case 20 duplication is compared with the three duplications (blue bars) and the deletion (red bar), involving SOX3 and its 5′ region previously reported in DSD patients,16, 17 based on UCSC Genome Browser (http://genome.ucsc.edu/), hg19.
The breakpoints of the duplications were finely mapped and cloned for both cases, (Figure 1b) and were located at 69 513 605 bps (proximally) and 69 692 812 bps (distally) for case 1 and at 69 404 081 bps (proximally) and 69 872 909 bps (distally) for case 2 (hg19), respectively. Three of the breakpoints occurred within regions of long interspersed nuclear elements, but there is no homology between the breakpoint sequences. Junction sequencing demonstrated that both duplications are direct. Case 1 junction shows insertion of four additional bases, probably derived by the duplication of adjacent sequence. Case 2 junction has a one-base overlap.
In case 3, karyotype analysis revealed the presence of a 46,XX karyotype with a de novo reciprocal translocation t(11;17)(p13;q24.3). Array-CGH analysis did not detect any imbalance but common CNVs, whereas FISH analysis allowed narrowing the breakpoints on the two derivative chromosomes. The BAC probe RP11-661B15 encompassed the breakpoint on chromosome 11, and the two BAC probes, CTD-2652P12 and RP11-879D6, encompassed the one on chromosome 17 (Figures 2a–b). Breakpoint mapping was performed by multiple long-range PCR reactions and sequencing, allowing to locate them at 35 935 981 bp on chromosome 11 and between 69 187 829 and 69 187 844 bp on chromosome 17, with a loss of 15 bp (Figure 2c). The rearrangement did not create or abrogate any predicted TFBS.
Translocation breakpoints mapping in case 3. (a) From the left: FISH analysis on patient’s lymphocytes with probes RP11-661B15 (chr11:35 875 774–36 048 561), RP11-879D6 (chr17:69 079 298–69 261 570), CTD-2652P12 (chr17:69 142 978–69 339 629), and a schematic representation of the results; arrowheads indicate the signals on the derivative chromosomes, whereas arrows mark normal chromosomes 11 and 17. (b) Map positions of the probes on chromosomes 11 and 17 highlighting the breakpoints (red arrowheads). (c) DNA sequences spanning the translocation breakpoints on der(11) and der(17) with the reference sequences.
Cases 4–19 did not show any significant CNV (Supplementary Table 1).
Case 20 showed a 46,XX karyotype. SNP-array analysis detected the presence of a 5.6 Mb duplication of the long arm of a chromosome X, involving the SOX3 gene (Figure 1c); the duplication was de novo and had occurred on the paternal allele. X-inactivation analysis showed a random pattern of inactivation.
Discussion
We present 19 unrelated cases of 46,XX subjects, with isolated abnormal gonadal development and male or ambiguous genitalia, all in the absence of the SRY gene. A further 46,XX SRY-negative subject showed syndromic DSD. A genomic imbalance or a chromosome rearrangement was detected in four. In three of them (cases 1, 3, and 20), external genitalia were unquestionably male with testes, whereas in one case (case 2) they were ambiguous with ovotestes.
Duplications of the desert region upstream of SOX9
Cases 1 and 2, an infertile male and a child with ovotestes, had partially overlapping duplications covering the so-called RevSex critical region on chromosome 17q24; such duplications have already been associated with 46,XX DSDs.4, 5, 6, 7 Our cases do not further narrow the minimal duplicated interval reported so far (Figure 3), but demonstrate that this genomic alteration is not rare among SRY-negative XX males and further establish that the duplication can be inherited by a healthy and fertile father. Although the frequency of genomic alterations involving the SOX9-coding region in 46,XX testicular or ovotesticular DSDs has been questioned,23 we identified copy number gains upstream of this gene in 2 of 19 novel cases with isolated 46,XX DSDs negative for SRY, either sporadic or familial, accounting for the 10.5% of our cohort. Considering that we have previously detected two further cases owing to the same cohort with similar RevSex duplications (Vetro et al,6 and a family that did not give consent to the publication), we may conclude that this frequency is even higher.
Overview of a 2.8-Mb screenshot of chromosome 17q24.3 (chr17:67 429 400–70 288 400, hg19) on the basis of UCSC Genome Browser. Copy-number variations upstream of SOX9 are shown. Blue bars: 46,XX DSD-associated duplications (DSD2 and DSD3 from Benko,4 and the cases reported by Cox,5 and Vetro,6 inherited the duplication from the healthy and fertile father; dark-green bars: duplications identified in brachydactyly-anonychia familial cases; red bars: 46,XY DSD-associated deletions (cases reported by Lecointre24 and Benko4 inherited the deletion from the mother; cases reported by Bhagavath25 inherited the deletion from the father); purple bars: deletions identified in patients with pathological phenotype not including DSD; light green: breakpoints of translocations identified in 46,XX subjects. The two cases with 46,XX DSDs (present report, case 3 and Refai15) are indicated by an asterisk. Breakpoints of balanced rearrangements, mainly associated with CD, ACD, or PRS are grouped into proximal (brown) and distal (dark-blue) clusters. The RevSex region is highlighted by a vertical light-orange bar. The hypothetical NGRE-containing region is depicted as a dashed blue line. References of all cases are provided on the left.4, 5, 6, 7, 11, 12, 13, 14, 15, 24, 25, 28, 29, 31, 32, 33, 34, 42, 43, 44, 45, 46 §: familial cases; PRS: Pierre-Robin sequence; ACD: acampomelic dysplasia CD: campomelic dysplasia; CHD: congenital heart defects.
In both our cases 1 and 2, the same duplication was present in the proband’s father, as for some previously reported cases, suggesting that a copy gain of the region does not affect sex development and fertility in 46,XY subjects, where SOX9 transcription is anyway activated during gonadal development. In a 46,XX background, in contrast, the presence of the duplication could be responsible of inappropriate expression of SOX9 in the embryo gonadal ridge.
Hypothetical mechanisms explaining the association between copy number gains at 17q24 and XX DSD are the following:
1. The duplication could alter SOX9 expression by increasing the dosage of one or more gonadal-specific enhancers located within the minimal duplicated interval defined as RevSex. This hypothesis is supported by the finding that deletions and duplications of the RevSex region have mirror effects, the firsts being associated with sex reversal in XY but not in XX subjects,4, 24 and the latter with XX sex reversal but no effect in XY individuals, as shown by familial cases4, 5, 6 (Figure 3). Therefore RevSex appears to be dosage sensitive although noteworthy exceptions, namely a duplication in one fertile XX female4 and a deletion in a fertile XY male,25 suggest that SOX9 dysregulation can, in some cases, be leaky possibly due to genomics modifier(s) of gonadal differentiation.
The minimal overlapping region of RevSex CNVs is ~70 kb (chr17:69 534 400–69 600 000, hg19). To explain why larger duplications containing RevSex are associated with brachydactyly-anonychia but not with sex reversal (dark green in Figure 3), we hypothesize that the additional copy of the critical region is placed too far from SOX9 to be able to influence its expression in the gonads.
2. An alternative model might consider that the amplifications abrogate SOX9 silencing by moving a hypothetical negative regulatory element located upstream of RevSex (negative gonadal regulatory element (NGRE), dashed blue line in Figure 3), too far away to exert any influence on SOX9 promoter. In fact, SOX9 repression needs to be maintained on the XX background both in the developing gonad26 and in the ovary,27 in order to ensure the differentiation and maintenance of ovarian cell fate. NGRE would not be displaced, but rather included in duplications associated with brachydactyly-anonychia but not with sex reversal (dark green in Figure 3). However according to this hypothesis, also proposed by Xiao et al,7 haploinsufficiency of such NGRE element should lead to the gonadal overexpression of SOX9, thus resulting in XX sex reversal. In contrast, a number of deletions, none of them associated with XX DSD, have been reported, covering the entire region delimited by the centromeric end of the RevSex duplications and the KCNJ2 gene. These individuals, either XX or XY, were investigated because of Pierre–Robin syndrome or cardiac defects.11, 13, 28, 29
The two duplications that we described are both in tandem, as reported for other DSD cases,4, 5 but a specific predisposing genomic architecture has not been highlighted. This duplication, although without effect in the XY background, appears to be very rare, with no cases containing at least the minimal duplicated RevSex region among the 14 316 individuals collected in the Database of Genomic Variants.30 This is in agreement with the extreme rarity of the SRY-negative 46,XX DSD condition.
Interruption of the desert region upstream of SOX9
We also report the second case of a balanced translocation associated with XX sex reversal (case 3). Breakpoint mapping allowed us to precisely define the 17q breakpoint of the t(11;17) translocation, which is located ~115 kb upstream of that reported by Refai et al15 The existence of a single gonadal-specific regulatory element interrupted by both these translocations is contradicted by the presence of translocation breakpoints in the same interval in at least three unrelated 46,XX females with normal sexual development (light green in Figure 3).31, 32, 33 As suggested,31 the chromatin environment of the recipient region may alter SOX9 regulation, even though in all these cases RevSex is translocated to the derivative chromosome together with SOX9, thus, in theory, retaining the cis-regulatory elements necessary for its gonadal expression.
Our patient 3 also shows signs of PRS. Several deletions and translocations, mapping in the region from 585 kb to 1.8 Mb upstream of SOX9, have been reported in patients with isolated PRS.11, 29, 31, 32, 34 These cases point to the existence of SOX9-regulatory elements, driving the expression of this gene in craniofacial structures, although no single element specifically impaired by the reported rearrangements has been yet identified.
SOX3 duplication
Finally, we report a new case of SOX3 duplication in a 46,XX boy ascertained because of mild developmental delay (case 20). SOX3, encoding a protein very similar to SRY, might be the ancestral SOX gene from which the SRY gene was derived.35
Duplications involving SOX3 and deletions of its 5′ region (Figure 1d) have been reported in at least four cases of XX DSD.16, 17 Moreover, a mouse model in which SOX3 is ectopically expressed in the developing gonads shows complete XX sex reversal, suggesting that gain-of-function mutations of SOX3 might act as an SRY surrogate in sex determination, promoting SOX9 gonadal expression.16 Interestingly, SOX3 duplications have been reported in two unrelated 46,XY individuals with X-linked hypopituitarism, whereas their carrier mothers were unaffected.36, 37 No hypopituitarism was present in our case 20 or in the patient reported by Moalem et al.17 An X-linked dominant but leaky mutation affecting sex development in a portion of XX subjects might be hypothesized, either as a consequence of the X-inactivation pattern in the developing gonad or of specific genomic modifiers.
Conclusions
We report additional evidences suggesting that, in the absence of SRY, altered expression of genes crucial to gonadal development, such as SOX9 and SOX3, may invert the expected embryonic plan.
Whereas for SOX3, it is easier to envisage a direct link between its duplication and increased gene expression,16 it is more difficult to understand the true functional link between duplications upstream of SOX9 and the different abnormal phenotypes, including gonadal abnormal differentiation.
Our study reports that the incidence for RevSex copy number gains associated with SRY-negative isolated 46,XX DSDs is >10%.
We can speculate that the RevSex duplication causes increased expression of SOX9 in undifferentiated gonadal cells, thus, resulting in testis differentiation even in the absence of SRY. In fact, duplications of SOX9 are associated with XX sex reversal not only in transgenic mice9 but also in the recently reported case of a deer, 38 and in three cases of dogs.39 Our case 3 shows that also interruption of the region upstream to the RevSex can result in XX sex reversal. Altogether our data reinforce the role of the desert region upstream of SOX9 in the regulation of this gene, as indicated by an altered histone methylation signature demonstrated in one of the RevSex duplicated cases.40 It is noteworthy that RevSex includes two lncRNAs, TCONS_00025195 and TCONS_00025196, with specific expression in the testis,41 possibly having a role in SOX9 transcriptional regulation.
References
Berta P, Hawkins JR, Sinclair AH et al: Genetic evidence equating SRY and the testis-determining factor. Nature 1990; 348: 448–450.
Jobling MA : A selective difference between human Y-chromosomal DNA haplotypes. Curr Biol 1998; 8: 4.
Rosser ZH, Balaresque P, Jobling MA : Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am J Hum Genet 2009; 85: 130–134.
Benko S, Gordon CT, Mallet D et al: Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet 2011; 48: 825–830.
Cox JJ, Willatt L, Homfray T, Woods CG : A SOX9 duplication and familial 46,XX developmental testicular disorder. N Engl J Med 2011; 364: 91–93.
Vetro A, Ciccone R, Giorda R et al: XX males SRY negative: a confirmed cause of infertility. J Med Genet 2011; 48: 710–712.
Xiao B, Ji X, Xing Y, Chen YW, Tao J : A rare case of 46, XX SRY-negative male with approximately 74-kb duplication in a region upstream of SOX9. Eur J Med Genet 2013; 56: 695–698.
Sekido R, Lovell-Badge R : Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453: 930–934.
Vidal VP, Chaboissier MC, de Rooij DG, Schedl A : Sox9 induces testis development in XX transgenic mice. Nat Genet 2001; 28: 216–217.
Huang B, Wang S, Ning Y, Lamb AN, Bartley J, Autosomal XX : sex reversal caused by duplication of SOX9. Am J Med Genet 1999; 87: 349–353.
Benko S, Fantes JA, Amiel J et al: Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet 2009; 41: 359–364.
White S, Ohnesorg T, Notini A et al: Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS One 2011; 6: e17793.
Sanchez-Castro M, Gordon CT, Petit F et al: Congenital Heart Defects in Patients with Deletions Upstream of SOX9. Hum Mutat 2013; 34: 1628–1631.
Kurth I, Klopocki E, Stricker S et al: Duplications of noncoding elements 5' of SOX9 are associated with brachydactyly-anonychia. Nat Genet 2009; 41: 862–863.
Refai O, Friedman A, Terry L et al: De novo 12;17 translocation upstream of SOX9 resulting in 46,XX testicular disorder of sex development. Am J Med Genet A 2010; 152A: 422–426.
Sutton E, Hughes J, White S et al: Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest 2011; 121: 328–341.
Moalem S, Babul-Hirji R, Stavropolous DJ et al: XX male sex reversal with genital abnormalities associated with a de novo SOX3 gene duplication. Am J Med Genet A 2012; 158A: 1759–1764.
Spielmann M, Klopocki E : CNVs of noncoding cis-regulatory elements in human disease. Curr Opin Genet Dev 2013; 23: 249–256.
Bonaglia MC, Giorda R, Beri S et al: Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet 2011; 7: e1002173.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW : Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239.
Rossi E, Giorda R, Bonaglia MC et al: De novo unbalanced translocations in Prader-Willi and Angelman syndrome might be the reciprocal product of inv dup(15)s. PLoS One 2012; 7: e39180.
Heinemeyer T, Wingender E, Reuter I et al: Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 1998; 26: 362–367.
Seeherunvong T, Ukarapong S, McElreavey K, Berkovitz GD, Perera EM : Duplication of SOX9 is not a common cause of 46,XX testicular or 46,XX ovotesticular DSD. J Pediatr Endocrinol Metab 2012; 25: 121–123.
Lecointre C, Pichon O, Hamel A et al: Familial acampomelic form of campomelic dysplasia caused by a 960 kb deletion upstream ofSOX9. Am J Med Genet Part A 2009; 149A: 1183–1189.
Bhagavath B, Layman LC, Ullmann R et al: Familial 46,XY sex reversal without campomelic dysplasia caused by a deletion upstream of the SOX9 gene. Mol Cell Endocrinol 2014; 393: 1–7.
Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B : Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet 2008; 17: 2949–2955.
Uhlenhaut NH, Jakob S, Anlag K et al: Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009; 139: 1130–1142.
Fukami M, Tsuchiya T, Takada S et al: Complex genomic rearrangement in the SOX9 5' region in a patient with Pierre Robin sequence and hypoplastic left scapula. Am J Med Genet A 2012; 158A: 1529–1534.
Gordon CT, Attanasio C, Bhatia S et al: Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence. Hum Mutat 2014; 35: 1011–1020.
Macdonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW : The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014; 42: D986–D992.
Fonseca AC, Bonaldi A, Bertola DR, Kim CA, Otto PA, Vianna-Morgante AM : The clinical impact of chromosomal rearrangements with breakpoints upstream of the SOX9 gene: two novel de novo balanced translocations associated with acampomelic campomelic dysplasia. BMC Med Genet 2013; 14: 50.
Hill-Harfe KL, Kaplan L, Stalker HJ et al: Fine mapping of chromosome 17 translocation breakpoints≥900 Kb upstream of SOX9 in acampomelic campomelic dysplasia and a mild, familial skeletal dysplasia. Am J Hum Genet 2005; 76: 663–671.
Velagaleti GV, Bien-Willner GA, Northup JK et al: Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet 2005; 76: 652–662.
Amarillo IE, Dipple KM, Quintero-Rivera F : Familial microdeletion of 17q24.3 upstream of SOX9 is associated with isolated Pierre Robin sequence due to position effect. Am J Med Genet A 2013; 161A: 1167–1172.
Foster JW, Graves JA : An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proc Natl Acad Sci U S A 1994; 91: 1927–1931.
Solomon NM, Nouri S, Warne GL, Lagerstrom-Fermer M, Forrest SM, Thomas PQ : Increased gene dosage at Xq26-q27 is associated with X-linked hypopituitarism. Genomics 2002; 79: 553–559.
Woods KS, Cundall M, Turton J et al: Over- and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet 2005; 76: 833–849.
Kropatsch R, Dekomien G, Akkad DA et al: SOX9 Duplication Linked to Intersex in Deer. PLoS One 2013; 8: e73734.
Rossi E, Radi O, De Lorenzi L et al: Sox9 duplications are a relevant cause of Sry-negative XX sex reversal dogs. PLoS One 2014; 9: e101244.
Lybaek H, de Bruijn D, den Engelsman-van Dijk AH et al: RevSex duplication-induced and sex-related differences in the SOX9 regulatory region chromatin landscape in human fibroblasts. Epigenetics 2014; 9: 416–427.
Smyk M, Szafranski P, Startek M, Gambin A, Stankiewicz P : Chromosome conformation capture-on-chip analysis of long-range cis-interactions of the SOX9 promoter. Chromosome Res 2013; 21: 781–788.
Blyth M, Huang S, Maloney V, Crolla JA, Karen Temple I : A 2.3 Mb deletion of 17q24.2-q24.3 associated with 'Carney Complex plus'. Eur J Med Genet 2008; 51: 672–678.
De Gregori M, Ciccone R, Magini P et al: Cryptic deletions are a common finding in "balanced" reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet 2007; 44: 750–762.
Jakubiczka S, Schroder C, Ullmann R et al: Translocation and deletion around SOX9 in a patient with acampomelic campomelic dysplasia and sex reversal. Sex Dev 2010; 4: 143–149.
Lestner JM, Ellis R, Canham N : Delineating the 17q24.2-q24.3 microdeletion syndrome phenotype. Eur J Med Genet 2012; 55: 700–704.
Pop R : Screening of the 1 Mb SOX9 5' control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet 2004; 41: e47–e47.
Acknowledgements
This work was supported by Telethon 2010 (GGP10121) and PRIN 2010-2011 (20108WT59Y_003) (both to O.) and by a grant of the Italian Ministry of Health (Ricerca Corrente 2013; to MC). We acknowledge the Galliera Genetic Bank member of ‘Network Telethon of Genetic Biobanks’ (project no. GTB12001A), funded by Telethon Italy, who provided us with specimen 01GGB13711M-13577. We would also acknowledge Dr Pietro Ameri (Internal Medicine, Genova University) for his collaboration.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Vetro, A., Dehghani, M., Kraoua, L. et al. Testis development in the absence of SRY: chromosomal rearrangements at SOX9 and SOX3. Eur J Hum Genet 23, 1025–1032 (2015). https://doi.org/10.1038/ejhg.2014.237
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.237
This article is cited by
-
Genetic control of typical and atypical sex development
Nature Reviews Urology (2023)
-
Duplication of SOX3 in an SRY-negative 46,XX male with prostatic utricle: case report and literature review
BMC Medical Genomics (2022)
-
Mammalian X-chromosome inactivation: proposed role in suppression of the male programme in genetic females
Journal of Genetics (2022)
-
Cellular fate of intersex differentiation
Cell Death & Disease (2021)
-
Mapping molecular pathways for embryonic Sertoli cells derivation based on differentiation model of mouse embryonic stem cells
Stem Cell Research & Therapy (2020)