Abstract
Bacterial chromosomes often carry integrated genetic elements (for example plasmids, transposons, prophages and islands) whose precise function and contribution to the evolutionary fitness of the host bacterium are unknown. The CTXφ prophage, which encodes cholera toxin in Vibrio cholerae1, is known to be adjacent to a chromosomally integrated element of unknown function termed the toxin-linked cryptic (TLC)2. Here we report the characterization of a TLC-related element that corresponds to the genome of a satellite filamentous phage (TLC-Knφ1), which uses the morphogenesis genes of another filamentous phage (fs2φ) to form infectious TLC-Knφ1 phage particles. The TLC-Knφ1 phage genome carries a sequence similar to the dif recombination sequence, which functions in chromosome dimer resolution using XerC and XerD recombinases3. The dif sequence is also exploited by lysogenic filamentous phages (for example CTXφ) for chromosomal integration of their genomes. Bacterial cells defective in the dimer resolution often show an aberrant filamentous cell morphology3,4. We found that acquisition and chromosomal integration of the TLC-Knφ1 genome restored a perfect dif site and normal morphology to V. cholerae wild-type and mutant strains with dif− filamentation phenotypes. Furthermore, lysogeny of a dif− non-toxigenic V. cholerae with TLC-Knφ1 promoted its subsequent toxigenic conversion through integration of CTXφ into the restored dif site. These results reveal a remarkable level of cooperative interactions between multiple filamentous phages in the emergence of the bacterial pathogen that causes cholera.
Similar content being viewed by others
Main
The TLC element of V. cholerae encodes the Cri replicase with homology to filamentous phage replication proteins and TlcR, a protein with sequence similarity to RstR, the repressor controlling lysogeny of the filamentous CTXφ and the target for anti-repression by the RstC product of satellite filamentous phage RS1φ (refs 1, 2, 5–9). For these reasons we speculated that the TLC element might correspond to the genome of a satellite filamentous phage that depended on another filamentous phage for its morphogenesis. As a prelude to the study described here, we devised a screen for the postulated ‘TLC helper phage’ and thus identified filamentous phage fs2φ as this helper10. In brief, our evidence (see Supplementary Information) that fs2φ is a TLC helper phage includes the following. First, strains encoding genetically marked versions of the TLC element (for example TLC-Kn1) inserted in their chromosome produce infectious TLC-Knφ1 phage particles only if also infected with fs2φ. Second, these TLC-related phage particles carry single-stranded DNA (ssDNA) corresponding to a circularized variant of the TLC element. Third, TLC-Knφ1 phages infect only cells expressing mannose-sensitive haemagglutinin (MSHA) pili, the known receptor of fs2φ (ref. 11). On infection of MSHA-positive vibrios, the TLC-Knφ1 ssDNA present in phage particles is converted to the double-stranded replicative form that is detectable in infected cells as a plasmid or as a chromosomally integrated copy. Fourth, the double-stranded replicative form of TLC-Knφ1 (designated pTLC-Kn1) was also shown to be sufficient for formation of TLC-Knφ1 phage in recipient cells provided that the cells are also infected with fs2φ. Thus, fs2φ is a helper phage that provides essential gene products required for morphogenesis of TLC-Knφ1 phage particles.
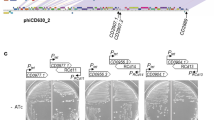
For a better understanding of the biology of TLCφ, we sequenced pTLC-Knφ1 and its chromosomally integrated form in strain AL33457-TLC-Kn1. Strain AL33457 was found to carry two copies of the TLC element that flank a unique open reading frame (ORF; VC1471; Fig. 1). Each of the two copies of chromosomally integrated TLC elements in AL33457 is composed of five ORFs, spanning from VC1466 to VC1470 and from VC1472 to VC1476, respectively. In strain AL33457-TLC-Kn1, the KnR determinant was located in VC1470 and thus, like ORF VC1471, it was located between the duplicated copies of TLC. Nucleotide sequence analysis of pTLC-Kn1 indicated that this plasmid probably formed as a result of recombination between two directly repeated 25-base-pair (bp) sequences (5′-ACATAATGCGCACTAGGAACATTTT-3′), which are located in the 3′ end of VC1465 and within VC1471 (Fig. 1). This 25-bp sequence within VC1471 overlaps by 18 bp (bold nucleotides) with the 28-bp dif1 sequence, 5′-ATTTAACATAACATACATAATGCGCACT-3′ (refs 12, 13). Dif1 is a site on the large chromosome of V. cholerae that is required for the XerC/XerD-mediated resolution of chromosome dimers, and similar sites are also exploited by various filamentous phages for integration of their genomes into the host chromosome using XerC/XerD-mediated recombination3,4,12,13,14,15. The dif1 sequence is used by CTXφ and RS1φ for their chromosomal integration though XerC/XerD-mediated recombination with the corresponding dif/attP site formed by the annealing of ssDNA derived from phage genomes12,13. The recombination event that formed pTLC-Kn1 looped out the entire region between the 25-bp duplicated sequence in VC1465 and VC1471, including the 18 bp identical to part of the dif1 sequence together with most of the ORF defined as VC1471. Thus, TLC-Knφ1 and pTLC-Kn1 encode part of the dif1 sequence (Fig. 1).
A conservative recombination event (dotted line and arrow) occurring between two identical 25-bp sequences (boxed nucleotides) located near the 3′ end of VC1465 and within VC1471 excised the plasmid from the chromosome of the V. cholerae AL33457-TLC-Kn1 parental strain. This strain carries two chromosomal copies of the TLC element with a kanamycin resistance (Kn) insert located in VC1470 of the first TLC element (TLC1). pTLC-Kn1 is therefore a circularized TLC-related element that carries most of VC1471 (a gene located between the 25-bp sequences). Within VC1471 there also exists an 18-bp region (red) that is identical to part of the known 28-bp dif1 sequence. TLC1 is composed of genes VC1466, VC1467, VC1468, VC1469 and VC1470; TCL2 is composed of genes VC1472, VC1473, VC1474, VC1475 and VC1476. For simplicity, only a subset of these genes is shown in this diagram.
These observations suggest that naturally occurring TLC-related phages might be capable of reconstituting a functional chromosomal dif sequence by recombining its partial dif site with a defective dif-like sequence during lysogenic integration of its genome into the chromosome. To test this hypothesis, we screened a collection of 97 clinical or environmental V. cholerae strains of both O1 and non-O1 serogroups and identified 18 strains that were negative for one or more chromosomal regions including TLC, VC1471 and the dif sequence3,12,13. These included 12 non-O1, non-O139 strains and 6 non-toxigenic O1 strains. Sequencing of the relevant region in five such TLC-negative CTX-negative O1 strains (AO12682, AO7543, AV2684248, AN19908 and AN25049; see Supplementary Table 1) revealed a gap between the rtxA gene (VC1451) and the gene designated VC1479 in all the five strains analysed. In toxigenic strains of the seventh pandemic El Tor biotype such as N16961, the CTX prophage and the TLC genes as well as the recombined dif-like sites formed by integration of CTXφ are located in this space16. In the intergenic region between rtxA and VC1479 in the TLC-negative strains we identified a possible defective dif-like sequence differing in two nucleotide pairs (G→A and C→T) from the genuine dif sequence (Fig. 2).
a, Schematic diagram showing site-specific integration of the TLC-Knφ1 genome into the chromosome of strain AO7543. The region in the vicinity of the phage attachment site is shown in green; the region in the vicinity of the chromosomal attachment site is shown in blue. The chromosomal attachment site corresponds to a defective dif-like sequence altered in an AT dinucleotide (arrows). Sequence analysis indicates that the recombination event that integrated the phage genome occurred in the central region of sequence identity (red), which also corresponds in part to the central region (CR) of a dif1 site (see b). This recombination event was probably generated at least in part by the action of XerC and XerD on chromosomal and phage nucleic acid substrates in that the TLC-Kn1 genome did not integrate chromosomally in XerC-defective and XerD-defective strains (see the text and Supplementary Information). Formation of the TLC prophage by integration resulted in the formation of a functional dif1 sequence (underlined) on its left border. The central sequence of identity is duplicated on the right border but the dif1 sequence is not duplicated. b, Base-pair alignments between the defective dif-like sequence on the chromosome of strain AO7543 that is the target for TLC-Knφ1 integration, the resultant hybrid sequence found after integration of TLC-Knφ1 (left end) and the authentic V. cholerae dif1 sequence. Colours of nucleotides correspond to those highlighted in a. The binding sequences for XerC and XerD recombinases are indicated by boxes.
Cultures of V. cholerae strains with deletions in the dif recombination site are known to contain a subpopulation of cells with a filamentous morphology3. These filaments reflect aberrant cell division resulting from a defect in XerC/XerD-mediated chromosome dimer resolution3. We examined whether naturally occurring V. cholerae O1 strains that lack TLC, or the genuine dif sequence, have a filamentous morphology. As shown in Fig. 3 and Supplementary Table 3, these strains do indeed have a filamentous morphology for a noticeable sub-population of their cells. We next tested whether transduction with TLC-Knφ1 phage could correct this morphology defect. In each case, cell filamentation in these V. cholerae strains was found to be eliminated after transduction with TLC-Knφ1 (Fig. 3 and Supplementary Table 3). These results suggest that these filamentous strains are indeed dif-deficient and that lysogeny with TLC-Knφ1 apparently corrected this defect.
a–c, Morphology of three strains (a, AO12682; b, AO7543; c, AV2684248) before infection with TLC-Knφ1; see Supplementary Tables 1 and 4 for details. d–f, Morphology of AO12682 (d), AO7543 (e) and AV2684248 (f) after infection and chromosomal integration of TLC-Knφ1.
For further verification of the natural formation of TLC-related phages and the role of the dif-like sequence encoded by VC1471 in the correction of dif-deficient phenotypes, we used a set of chromosomal transposon insertion mutants of C6706 (ref. 17). We selected five strains carrying TnFGL3 insertions in the different ORFs of TLC (VC1466, VC1467, VC1468, VC1469, VC1470 and VC1471). With the exception of VC1469, for each strain we were able to recover a plasmid corresponding to the double-stranded replicative form of the TLCφ-related genome with a TnFGL3 insertion in the corresponding ORF. No plasmid recovery was actually expected for the insertion mutant in VC1469 because this gene encoded Cri, the protein required for the replication of TLC-related plasmids2. When these TLC-related plasmids were introduced into each of the strains SA317, AO7543 and AO12682 (naturally occurring strains with a filamentous morphology and negative for VC1471), normal cellular morphology was restored except in one case. The double-stranded replicative-form plasmid derived from the mutant carrying a TnFGL3 insertion in VC1471 failed to complement the morphology defect in these strains (Supplementary Fig. 4). The TnFGL3 insertion in VC1471 is located within the dif-like sequence (bold) encoded by VC1471 (insertion indicated by the asterisk in the sequence 5′-ACATACA*TAATGCGCACTAGGAACA-3′). We conclude that the dif-like sequence present in VC1471 is required to correct morphological defects when TLC-related plasmids are introduced into naturally occurring dif−/TLC − strains of V. cholerae.
We studied the integration of the TLC-Knφ1 genome into the chromosome of the naturally occurring TLC-negative strains AO7543 and AO12682 after infecting these strains with TLC-Knφ1 phage particles. As expected, Southern blot hybridization (Supplementary Fig. 5) and polymerase chain reaction (PCR) analysis indicated that the TLC-Knφ1 genome had inserted into the bacterial genome in the intergenic region between rtxA and VC1479. The correction of the cell filamentation phenotype of dif-deficient strains was observed only in strains in which the TLC-Knφ1 genome had integrated into the chromosome. In contrast, introduction of a pUC18 clone of VC1471, designated pVC1471, into the defective cells did not cure the cell filamentation phenotype even though it carried the dif-like sequence. This finding indicated that the dif-like sequence present in VC1471 functions only in cis and not in trans. This is what one would expect if the dif-like sequence in VC1471 could indeed function in recombination with XerC/XerD to resolve chromosomal dimers only if it recombined into the chromosome. To verify this assumption, we used mutants of dif− strains with transposon insertions in the XerC or XerD genes. As expected, transduction of TLC-Knφ1 into these XerC-defective or XerD-defective strains did not cure their cell filamentation phenotype (Supplementary Table 4). Furthermore, PCR-based analyses as described above confirmed that the TLC-Knφ1 DNA did not integrate into the chromosome of the XerC or XerD mutant strains.
To further examine the mechanism associated with the elimination of the dif− defect through the chromosomal integration of TLC-Knφ1 DNA, we sequenced the junction of several independent TLC-Knφ1 integration events. The sequence analysis showed that TLC-Knφ1 DNA had integrated into the intergenic region between rtxA and VC1479 in strains AO7543, AV2684248 and AO12682 by using its defective dif-like site as a recombinational substrate (Fig. 2a). The recombination event leading to the integration of the TLC-Knφ1 genome resulted in the formation of a functional dif sequence identical to the genuine dif1 sequence reported for V. cholerae3,13 (Fig. 2b). This result also suggests that TLC-deficient V. cholerae strains contain alternative dif-like sequences that can still function in recombination with a TLC-encoded dif-like sequence but are not fully functional in chromosome dimer resolution. We conclude that the dif-like site in VC1471 recombines with the defective chromosomal dif-like sequence in these TLC-negative strains during the process of XerC/D-mediated TLC genome insertion and that the product of this integration event generates a dif sequence that is functional in chromosomal dimer resolution.
In toxigenic V. cholerae, the CTXφ genome exists as a prophage inserted into the bacterial chromosome at the dif recombination site12,13,15. Because transduction with TLC-Knφ1 reconstructs a functional dif sequence in the recipient bacterium (Fig. 2), we tested whether TLC-Knφ1 transductants could be stably lysogenized by CTXφ. We chose test strains that were positive for the TCP locus (which encodes the receptor for CTXφ (ref. 1)), and used a CTXφ prophage that was marked with a chloramphenicol resistance marker (CTX-Cmφ). As expected, we found that TLC-Knφ1 transductants were readily superinfected with CTX-Cmφ and in these cases CTX-Cmφ was found integrated into the dif site generated through previous integration of TLC-Knφ1. In contrast, although natural TLC-negative strains could also be infected with CTX-Cmφ (Supplementary Table 6), the CTX-Cmφ genome did not integrate, and the unintegrated CTX-Cmφ genome was rapidly lost when inoculated into the intestinal loops of adult rabbits. Because the integrated form of the CTXφ genome is known to be more stably retained in V. cholerae than the un-integrated plasmid form is18, these data strongly argue that TLCφ is crucial to the natural, stable acquisition of CTXφ.
Although the TLC element was known to be invariably present in all CTX-positive strains and notably absent in CTX-negative strains2, the role of this element in the evolution of toxigenic V. cholerae was not clear until the present study. Here we show that the TLC element can give rise to infectious phage particles (TLCφ) when its morphogenesis is supported by another filamentous phage, previously designated fs2φ (ref. 10). Furthermore, infectious forms of TLCφ that encode a dif-like sequence can be easily isolated. These specialized TLCφ-related transducing phages can, after chromosomal integration, generate a functional dif sequence and correct aberrant filamentous morphology present in TLC-negative cells that apparently show defective dif/XerC/XerD-mediated chromosome dimer resolution. Lysogeny by theae TLC phages leads to the restoration of a functional dif site, which is also essential for the stable integration of CTXφ and the conversion of V. cholerae to a toxigenic form.
The most common strains of V. cholerae causing cholera in the world today are all highly related to the seventh pandemic clone of V. cholerae, which emerged as a human pathogen in 1971 in the Celebes Islands19,20. The arrangement of the TLC prophage, and the dif site used by CTXφ in these highly successful pandemic strains, is virtually the same as the one that we produced experimentally in this study by lysogeny of dif− strains such as AO7543 sequentially with TLC-Knφ1 followed by CTX-Cmφ. It therefore seems highly likely that the precursor of the seventh pandemic clone was a dif− strain that emerged as a pandemic pathogen after sequential lysogeny by three filamentous phages, namely TLCφ, CTXφ and RS1φ (Fig. 4). Because dif− defects are deleterious to growth, it is possible that the precursor of the seventh pandemic clone may be rare in the environment or that the dif− genotype confers an as yet undetermined advantage for non-toxigenic O1 strains in the context of its environmental niche. Nonetheless, our data suggest that the evolutionary emergence of the toxigenic seventh pandemic clone of V. cholerae probably involved molecular interactions between two satellite filamentous phages (TLCφ and RS1φ), three helper filamentous phages (fs2φ, CTXφ and KSFφ (ref. 21)), and two type IV pilus-based phage receptors (MSHA and TCP) (Supplementary Fig. 6). Accordingly, our results provide a model for understanding the cooperative interactions of multiple genetic elements in the evolution of pathogenic bacteria from non-pathogenic environmental progenitors.
Boxes indicate the integrated prophages and dif sequence in the seventh pandemic strain, and the solid lines indicate the corresponding empty sites where these elements are absent in the precursor strains. Sequential lysogeny by three different filamentous phages (TLCφ, CTXφ and RS1φ) and the role of two helper phages (fs2 and KSF-1) are shown. To generate the observed organization of RS1 prophages found in most seventh pandemic strains, it is postulated that two rounds of RS1 prophage integration would be needed if this prophage integrated into only a functional dif site and then reconstituted only one dif site after integration. For a more complete explanation of these hypothetical steps see Supplementary Fig. 6.
Methods Summary
A genetic marker encoding kanamycin resistance (KnR) was introduced into the TLC element carried by multiple V. cholerae strains followed by screening of the marked strains for the production of TLC-related KnR transducing particles in the culture supernatants. Digestion with mung bean nuclease and also hybridization analysis with strand-specific oligonucleotide probes corresponding to the (+) and (−) strands of the TLC element were conducted to test whether the DNA carried by putative TLC-related phage particles present in filter-sterilized culture supernatants was single-stranded. The role of fs2φ as a helper of TLC satellite phage was established by demonstrating the formation of TLC-Knφ1 phage in recipient cells that contained pTLC-Kn1, provided that the cells were also infected with fs2φ.
For transduction assays, recipient V. cholerae strains were mixed with genetically marked phage preparations5, and transductants were selected by using Luria–Bertani agar medium containing appropriate antibiotics. Integration of the TLC-Knφ1 genome was detected by Southern blot hybridization and PCR assays using two primers, of which one was complementary to the chromosomal region and the other corresponded to pTLC-Knφ1. DNA sequencing was conducted for further confirmation of the integration event and to detect the generation of the dif sequence. Subsequently, a chloramphenicol resistance (CmR)-marked CTX phage was used to study the susceptibility and chromosomal integration5,18 of CTX phages into the restored dif site. The CmR-marked CTX phage genome (pCTX-Cm) was constructed by replacing the KnR marker in pCTX-Km (ref. 1) derived from strain SM44 with a CmR cassette.
The full list of strains and plasmids is available as Supplementary Table 1. Full methods and associated references are provided in Supplementary Methods.
References
Waldor, M. K. & Mekalanos, J. J. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science 272, 1910–1914 (1996)
Rubin, E. J., Lin, W., Mekalanos, J. J. & Waldor, M. K. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28, 1247–1254 (1998)
Huber, K. E. & Waldor, M. K. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417, 656–659 (2002)
Blakely, G. et al. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75, 351–361 (1993)
Faruque, S. M. et al. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXφ. Infect. Immun. 70, 163–170 (2002)
Davis, B. M., Kimsey, H. H., Kane, A. V. & Waldor, M. K. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21, 4240–4249 (2002)
Kimsey, H. H. & Waldor, M. K. The CTXφ repressor RstR binds DNA cooperatively to form tetrameric repressor-operator complexes. J. Biol. Chem. 279, 2640–2647 (2004)
Waldor, M. K., Rubin, E. J., Pearson, G. D., Kimsey, H. & Mekalanos, J. J. Regulation, replication, and integration functions of the Vibrio cholerae CTXΦ are encoded by region RS2. Mol. Microbiol. 24, 917–926 (1997)
Davis, B. M. & Waldor, M. K. CTXΦ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl Acad. Sci. USA 97, 8572–8577 (2000)
Ikema, M. & Honma, Y. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144, 1901–1906 (1998)
Ehara, M. et al. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol. Lett. 154, 293–301 (1997)
Val, M. E. et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae . Mol. Cell 19, 559–566 (2005)
Das, B., Bischerour, J., Val, M. E. & Barre, F. X. Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl Acad. Sci. USA 107, 4377–4382 (2010)
Iida, T. et al. Filamentous bacteriophage of Vibrios are integrated into the dif-like site of the host chromosome. J. Bacteriol. 184, 4933–4935 (2002)
McLeod, S. M. & Waldor, M. K. Characterization of XerC and XerD-dependent CTX phage integration in Vibrio cholerae . Mol. Microbiol. 54, 935–947 (2004)
Heidelberg, J. F. et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae . Nature 406, 477–483 (2000)
Cameron, D. E., Urbach, J. M. & Mekalanos, J. J. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae . Proc. Natl Acad. Sci. USA 105, 8736–8741 (2008)
Faruque, S. M. et al. Diminished diarrheal response to Vibrio cholerae strains carrying the replicative form of the CTXΦ genome instead of CTXΦ lysogens in adult rabbits. Infect. Immun. 69, 6084–6090 (2001)
Faruque, S. M., Albert, M. J. & Mekalanos, J. J. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae . Microbiol. Mol. Biol. Rev. 62, 1301–1314 (1998)
Kaper, J. B., Morris, J. G. & Levine, M. M. Cholera. Clin. Microbiol. Rev. 8, 48–86 (1995)
Faruque, S. M. et al. CTX phage-independent production of RS1 satellite phage by Vibrio cholerae . Proc. Natl Acad. Sci. USA 100, 1280–1285 (2003)
Acknowledgements
We thank T. M. Zaved Waise and S. M. Nashir Udden for technical assistance. This research was funded in part by the National Institutes of Health (grants RO1-GM068851 and RO1-AI070963) under different sub-agreements between the Harvard Medical School and the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B). The ICDDR,B is supported by countries and agencies that share its concern for the health problems of developing countries.
Author information
Authors and Affiliations
Contributions
F.H., M.K. and S.M.F. conducted the experiments and performed analyses of bacterial strains and phages. S.M.F. and J.J.M. designed the studies, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, additional references, Supplementary Tables 1-6 and Supplementary Figures 1- 6 with legends. (PDF 2619 kb)
Rights and permissions
About this article
Cite this article
Hassan, F., Kamruzzaman, M., Mekalanos, J. et al. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467, 982–985 (2010). https://doi.org/10.1038/nature09469
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09469
This article is cited by
-
Comparative genomics of 274 Vibrio cholerae genomes reveals mobile functions structuring three niche dimensions
BMC Genomics (2014)
-
Holliday junction affinity of the base excision repair factor Endo III contributes to cholera toxin phage integration
The EMBO Journal (2012)
-
A hybrid approach for the automated finishing of bacterial genomes
Nature Biotechnology (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.