Abstract
Higher eukaryotic genomes are bound by a large number of coding and non-coding RNAs, but approaches to comprehensively map the identity and binding sites of these RNAs are lacking. Here we report a method to capture in situ global RNA interactions with DNA by deep sequencing (GRID-seq), which enables the comprehensive identification of the entire repertoire of chromatin-interacting RNAs and their respective binding sites. In human, mouse, and Drosophila cells, we detected a large set of tissue-specific coding and non-coding RNAs that are bound to active promoters and enhancers, especially super-enhancers. Assuming that most mRNA–chromatin interactions indicate the physical proximity of a promoter and an enhancer, we constructed a three-dimensional global connectivity map of promoters and enhancers, revealing transcription-activity-linked genomic interactions in the nucleus.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Fu, X.D. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 1, 190–204 (2014).
Rinn, J.L. & Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
West, J.A. et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 (2014).
Chu, C., Qu, K., Zhong, F.L., Artandi, S.E. & Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol. Cell 44, 667–678 (2011).
Simon, M.D. et al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 108, 20497–20502 (2011).
Engreitz, J.M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Fullwood, M.J. et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Zhang, Y. et al. Chromatin connectivity maps reveal dynamic promoter–enhancer long-range associations. Nature 504, 306–310 (2013).
Dixon, J.R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Whyte, W.A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Pott, S. & Lieb, J.D. What are super-enhancers? Nat. Genet. 47, 8–12 (2015).
Grosswendt, S. et al. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell 54, 1042–1054 (2014).
Engreitz, J.M. et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199 (2014).
Chen, L.L. & Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478 (2009).
Yaffe, E. & Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 (2011).
Gelbart, M.E. & Kuroda, M.I. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136, 1399–1410 (2009).
Alekseyenko, A.A. et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609 (2008).
Straub, T., Grimaud, C., Gilfillan, G.D., Mitterweger, A. & Becker, P.B. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 4, e1000302 (2008).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Gallo, S.M. et al. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 39, D118–D123 (2011).
Nègre, N. et al. A cis-regulatory map of the Drosophila genome. Nature 471, 527–531 (2011).
Adelman, K. & Lis, J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012).
Giorgetti, L. et al. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016).
Ulianov, S.V. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26, 70–84 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Di Stefano, M., Paulsen, J., Lien, T.G., Hovig, E. & Micheletti, C. Hi-C-constrained physical models of human chromosomes recover functionally-related properties of genome organization. Sci. Rep. 6, 35985 (2016).
Schmitz, K.M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Sigova, A.A. et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015).
Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013).
Wei, C. et al. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol. Cell 62, 875–889 (2016).
Yin, Y. et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516 (2015).
Lee, N., Moss, W.N., Yario, T.A. & Steitz, J.A. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160, 607–618 (2015).
Yu, Y. et al. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339–342 (2015).
Core, L.J., Waterfall, J.J. & Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Servant, N. et al. HiTC: exploration of high-throughput 'C' experiments. Bioinformatics 28, 2843–2844 (2012).
Phanstiel, D.H., Boyle, A.P., Araya, C.L. & Snyder, M.P. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810 (2014).
Core, L.J. et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 (2012).
Acknowledgements
We wish to express our gratitude to X. Ji and R. Young (Massachusetts Institute of Technology) for sending us MM.1S cells; B. Zhou and S. Wasserman (University of California San Diego) for sending us S2 cells; G. Li and B. Ren (University of California San Diego) for sharing mESC with us; C.-M. Chiang (University of Texas Southwestern) for sharing the JQ1 inhibitor; C. Class, S. Dowdy, and N. Chi for critical comments; and members of the Fu laboratory for stimulating discussion and advice during this investigation. This work was supported by NIH grants (HG004659, HG007005, GM049369 and DK098808) to X.-D.F.
Author information
Authors and Affiliations
Contributions
X.L. and X.-D.F. designed GRID-seq; X.L. performed most experiments; B.Z. and X.L. analyzed the data; L.C. performed GRO-seq; L.-T.G. contributed to characterization of the global gene network; H.L. sequenced all GRO-seq and GRID-seq libraries; X.L., B.Z., and X.-D.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Characterization of the GRID-seq technology.
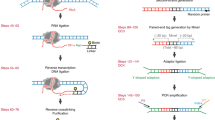
a, The design of a bivalent linker for GRID-seq. The top strand is a 5’ phosphorylated DNA sequence (black) and the bottom strand consists of both DNA and RNA bases (purple) with a biotinylated T residue (red) in the middle. Randomized bases (N) served as barcodes for filtering PCR duplicates during library amplification and both ends of the linker each carry an MmeI restriction site (grey-shaded). The linker is pre-adenylated for ligation to RNA in the absence of ATP, which prevents ligation of endogenous RNAs. b, Characterization of the linker before (left) and after (right) annealing, showing the sensitivity of the RNA-containing linker to RNase A. c, Controls by omitting RNA ligase, DNA ligase or both during GRID-seq library construction, which yielded expected ligated products of singleton tags or paired-end tags (left). After adapter ligation (middle) and PCR amplification (right), the expected products were only detected after library construction with both RNA and DNA ligases.
Supplementary Figure 2 Characterization of GRID-seq libraries.
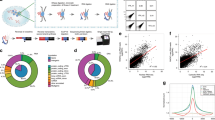
a, Summary of sequenced GRID-seq libraries constructed on two human cell lines (MDA-MB-231 and MM.1S), one mESC, and one Drosophila S2 cells. Shown are raw reads, linker-containing reads, and uniquely mapped reads from mated RNA/DNA pairs. b, Nucleotide frequency of DNA (up) and RNA (bottom) reads. Note specific dinucleotide as part of the AluI recognition site at the 3’ end of DNA reads, but the lack of nucleotide bias in any position of RNA reads. c,d, Strand orientation of mapped RNA (c) and DNA (d) reads. Note the same strand orientation of mapped RNA reads as their transcripts, but not DNA. e,f, Reproducibility of GRID-seq libraries constructed on human (e) and Drosophila (f) cells. g,h, Comparison of GRID-seq detected RNA reads with gene expression detected by RNA-seq of rRNA-depleted total RNA (g) or GRO-seq (h) in Drosophila S2 cells. The lncRNA roX2 is highlighted in both plots. RPK: GRID-seq reads per Kb. RPKM: reads per Kb per million mapped reads.
Supplementary Figure 3 Characteristics of MALAT1 interaction with chromatin in human and mouse cells.
a, MDA-MB-231 cells (upper panel) and mESCs (bottom panel). b, Meta-analysis of MALAT1-chromatin interactions in human (left panel) and mouse (right panel) cells on a composite gene model. TSS: Transcription Start Site. TTS: Transcription Termination Site. c, Comparison of MALAT1-chromatin interactions detected by GRID-seq versus RAP-DNA in mESCs. d, Comparison of MALAT1-chromatin interactions detected by GRID-seq in MDA-MB-231 cells versus those identified by CHART in MCF-7 cells. Note that these meta-analysis plots were based on the data before background correction, which remained the same after background subtraction. P-values in a, c, and d were determined by Fisher’s exact test.
Supplementary Figure 4 Construction of a background model for GRID-seq data analysis.
a, Steps in building the background with trans-chromosomal interacting mRNAs. GRID-seq detected raw DNA reads of trans-acting mRNAs (n=3,559) on chromatin were displayed on all chromosomes with their cis-acting signals masked by grey boxes. Two specific mRNAs on their trans locations were highlighted below the heatmap. Step 1: All trans-acting mRNAs were summed, and then in Step 2, normalized to 1 million total reads. Step 3: The GRID-seq signals of the two specific mRNAs were similarly normalized to 1 million across all chromosomes. Red signals above the general background were highlighted. Step 4: Specific interaction sites were displayed after background correction. b, Comparison of deduced background from replicate GRID-seq libraries constructed on human MDA-MB-231 cells, mESCs and Drosophila S2 cells. Each genome was 1 Kb binned for global comparison. RPK: reads per Kb. c, Comparison between the true background based on the human-Drosophila mixing experiment and the deduced background from mRNA trans-chromosomal reads in inferring specific RNA-chromatin interactions. The data illustrate <1% discrepancies in inferring cis, trans, or all reads in both MDA-MB-231 and S2 datasets. d, Quantification of cis and trans-chromosomal reads distribution before and after background correction. e,f,g, Background GRID-seq signals in open chromatin regions in human (e), mouse (f), and Drosophila (g) cells. Chromatin marks and RNA Pol II binding were displayed on autoscale.
Supplementary Figure 5 Characteristics of RNA-chromatin interactions.
a, RNA-chromatin interaction heatmaps on Chr. 5 constructed from two independent GRID-seq libraries on MDA-MB-231 cells. Boxed regions in each panel were enlarged with increasing resolution in the next panel on the right. A set of representative chromatin-enriched RNAs are labeled on the right and their encoding gene loci illustrated on the bottom. b, Pearson’s Correlation Coefficient of GRID-seq interaction density of each RNA at increasing resolution (decreasing bin size) across genomes between the replicates performed on MDA-MB-231 cells, mESC and Drosophila S2 cells. c, Whole chromosome view of raw roX2 GRID-seq DNA reads on Drosophila Chr. X and Chr. 3R in comparison with raw signals detected by ChIRP and CHART, as well as ChIP-seq signals for the roX2 binding protein MSL3. d, Percentage of total raw roX2 GRID-seq DNA reads on Chr. X in comparison with the raw data of ChIRP, CHART, GRID-seq and MSL3 ChIP-seq. Total number of raw read for each dataset is noted on top of each bar. e, Comparison of roX2 peaks identified by ChIRP, CHART, and GRID-seq with MSL3 ChIP-seq peaks in Drosophila S2 cells, showing the highest overlap between GRID-seq and MSL3 ChIP-seq identified peaks. f, Comparison of GRID-seq identified roX2 peaks with MSL3 ChIP-seq peaks on Chr. X and autosomes, showing their overlap only on Chr. X. g, Meta-analysis of roX2 GRID-seq DNA reads relative to chromatin binding of CES and HAS. Pie charts on the right illustrate the overlaps between CES/HAS peaks and roX2 GRID-seq peaks. h,i,j, Circos plots depicting chromatin-interactions of two representing lncRNAs (left panels) and two protein-coding mRNAs (right panels) in each cell type. In MDA-MB-231 cells (h), the lncRNA MALAT1 broadly interacted with all chromosomal regions whereas PVT1 was predominantly involved in local and cis-chromosomal interactions, although trans-chromosomal interaction signals were also evident in various specific locations (h, left panel). Similarly, RNAs from the coding HMGA2 gene showed chromatin interactions in local, cis and trans modes whereas RNAs from the coding SMAD5 gene was largely confined in local interactions (h, right panel). Similar examples also illustrated distinct chromatin interaction patterns with both coding RNAs and lncRNAs in mESC (i) and Drosophila S2 cells (j).
Supplementary Figure 6 Global view of RNA-chromatin interactions in human MM.1S cells.
a, Heatmap showing RNA-chromatin interactions across the whole human genome in MM.1S cells. Row: chromatin-enriched RNAs. Column: human genome in 1 Mb resolution. Representative trans-chromosomal interacting RNAs are labeled on the right. b, Enlarged heatmaps of boxed Chr. 22 and X in a, showing detailed RNA-chromatin interactions on Chr. 22 (left) and Chr. X (right). Representative RNAs are labeled on the left (pc: protein-coding RNAs, nc: non-coding RNAs), highlighting the non-coding RNA XIST on Chr. X (note that XIST is expressed in MM.1S cells, but not in MDA-MB-231 cells). Top: The background deduced from endogenous traveling mRNAs.
Supplementary Figure 7 RNA-chromatin interactions on cell type-specific enhancers.
a,b, Scatterplots of foreground (a) and background (b) of GRID-seq signals between MDA-MB-231 and MM.1S cells in the 1 Kb-binned human genome. c,d, Violin plots showing co-enrichment of specific RNA-chromatin interactions and key chromatin features in MDA-MB-231 cells (c) and MM.1S (d). Left: Enrichment of mean chromatin interaction signals of GRID-seq RNA density after background correction relative to ChIP-seq peaks of RNA Pol II, H3K4me1, H3K4me3, H3K27ac and H3K27me3. Right: Background signals. RPK: reads per Kb. Bars represent the range from 25 to 75 percentiles. e,f, Quantification of RNA-chromatin interaction levels on enhancers in the same cell type (orange) relative to specific enhancers in a different cell type (grey). g, Correlation of RNA-chromatin interaction levels on promoters with gene expression in MDA-MB-231 and MM.1S cells.
Supplementary Figure 8 RNA interaction levels on super-enhancers and relative contribution of typical and super-enhancers to transcription.
a, Super-enhancers in relationship to RNA-chromatin interaction levels. Enhancers in MDA-MB-231 cells were ranked by RNA interaction levels. Each red bar on top represents a super-enhancer. Red curve: The accumulative curve of rank-ordered RNA-chromatin interactions; Grey dashed line: Random distribution. b, Density distribution of chromatin-enriched RNAs at super-enhancers (SE, red) and typical enhancers (TE, blue). c, Left: Rank-ordered RNA-chromatin interaction levels at all active enhancers. Right: The Top 10% and bottom 10% enhancers with the most and the least RNA interactions, respectively, selected for functional analysis. d, Gene transcription associated with top 10% enhancers (orange box) and bottom 10% enhancers (green box). e, Accumulation curves of transcription changes of genes associated with top 10% and bottom 10% enhancers in response to JQ1 treatment in MDA-MB-231 cells. Statistical significance of comparison is estimated by Student’s t-test in panel b, d and e.
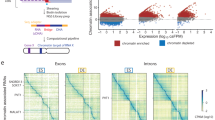
Supplementary Figure 9 Comparison between Hi-C and GRID-seq in different cell types.
a, Examples of Hi-C interactions in comparison with GRID-seq detected RNA interactions relative to previously assigned TADs in Drosophila S2 cells. H3K27ac ChIP-chip signals and reporter scored enhancers from the RedFly database were displayed at bottom. b,c, Left panels: GRID-seq signals relative established TADs in mESCs (b) and Drosophila S2 cells (c). Right panels: GRID-seq signals across TAD boundaries. Red lines indicate chromatin-enriched RNAs with more than 50% GRID-seq signals spread to adjacent TADs.
Supplementary Figure 10 Inferred enhancer-promoter distance and examples.
a, Density distributions of cis- and trans-chromosomal RNA interaction levels on enhancers in MM.1S cells. x-axis: Z-scores of all trans-chromosomal RNA interactions (green). Z≥3 was used to identify significant RNA interactions on individual promoter and enhancer elements. b,c, Distribution of linear DNA distance between genes and RNA decorated super-enhancers (b) and typical enhancers (c). d, Circos plot, showing a representative case of two chromatin-enriched RNAs RRBP1 and SNX5 on nearby seven enhancers, one of which corresponds to a super-enhancer (red) in MM.1S cells. The RRBP1 RNA interaction profile is shown on the outer track (green) and the SNX5 RNA interaction profile on the inter track (blue). Ribbons connecting with enhancers illustrate inferred enhancer-promoter association. e, f, Upon JQ1 treatment, fold-changes in gene expression are shown in e and fold-changes in BRD4 binding on individual enhancers in f.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 1747 kb)
Supplementary Table 1
GRID-seq Enriched RNA on Chromatin from Two Human Cell Lines, One Mouse ES Cell Line, and One Drosophila Cell Line (XLSX 385 kb)
Supplementary Table 2
GRID-seq Deduced Promoter-Enhancer Interactions in MM.1S Cells (XLSX 1023 kb)
Supplementary Table 3
Public Datasets Used for Comparison and Cross-Analysis in the Current Study (XLSX 48 kb)
Rights and permissions
About this article
Cite this article
Li, X., Zhou, B., Chen, L. et al. GRID-seq reveals the global RNA–chromatin interactome. Nat Biotechnol 35, 940–950 (2017). https://doi.org/10.1038/nbt.3968
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3968