Abstract
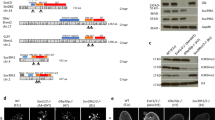
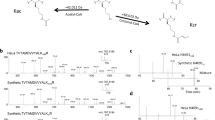
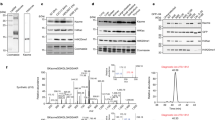
We report the identification of a new type of histone mark, lysine 2-hydroxyisobutyrylation (Khib), and identify the mark at 63 human and mouse histone Khib sites, including 27 unique lysine sites that are not known to be modified by lysine acetylation (Kac) and lysine crotonylation (Kcr). This histone mark was initially identified by MS and then validated by chemical and biochemical methods. Histone Khib shows distinct genomic distributions from histone Kac or histone Kcr during male germ cell differentiation. Using chromatin immunoprecipitation sequencing, gene expression analysis and immunodetection, we show that in male germ cells, H4K8hib is associated with active gene transcription in meiotic and post-meiotic cells. In addition, H4K8ac-associated genes are included in and constitute only a subfraction of H4K8hib-labeled genes. The histone Khib mark is conserved and widely distributed, has high stoichiometry and induces a large structural change. These findings suggest its critical role on the regulation of chromatin functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chi, P., Allis, C.D. & Wang, G.G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Berger, S.L. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Heintzman, N.D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Montellier, E., Rousseaux, S., Zhao, Y. & Khochbin, S. Histone crotonylation specifically marks the haploid male germ cell gene expression program post-meiotic male-specific gene expression. Bioessays 34, 187–193 (2012).
Montellier, E. et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 27, 1680–1692 (2013).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 (2012).
Chen, Y. et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 (2007).
Chen, Y., Chen, W., Cobb, M.H. & Zhao, Y. PTMap—a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl. Acad. Sci. USA 106, 761–766 (2009).
Rohwerder, T. & Mueller, R.H. Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb. Cell Fact. 9, 13 (2010).
Kumps, A., Duez, P. & Mardens, Y. Metabolic, nutritional, iatrogenic, and artifactual sources of urinary organic acids: a comprehensive table. Clin. Chem. 48, 708–717 (2002).
Freitas, M.A., Sklenar, A.R. & Parthun, M.R. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J. Cell. Biochem. 92, 691–700 (2004).
Wisniewski, J.R., Zougman, A. & Mann, M. Nɛ formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 36, 570–577 (2008).
Olsen, J.V. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling 3, ra3 (2010).
Drogaris, P. et al. Histone deacetylase inhibitors globally enhance H3/H4 tail acetylation without affecting H3 lysine 56 acetylation. Sci. Rep. 2, 220 (2012).
Gorovsky, M.A. et al. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb. Symp. Quant. Biol. 42, 493–503 (1978).
Morinière, J. et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664–668 (2009).
Gaucher, J. et al. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 31, 3809–3820 (2012).
Turner, J.M.A. Meiotic sex chromosome inactivation. Development 134, 1823–1831 (2007).
Namekawa, S.H. et al. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16, 660–667 (2006).
Reynard, L.N. & Turner, J.M.A. Increased sex chromosome expression and epigenetic abnormalities in spermatids from male mice with Y chromosome deletions. J. Cell Sci. 122, 4239–4248 (2009).
Maxwell, P.H. et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 (1999).
Katada, S., Imhof, A. & Sassone-Corsi, P. Connecting threads: epigenetics and metabolism. Cell 148, 24–28 (2012).
Kaelin, W.G. & McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013).
Garcia, B.A. et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2, 933–938 (2007).
Pivot-Pajot, C. et al. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol. Cell. Biol. 23, 5354–5365 (2003).
Kotaja, N. et al. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat. Methods 1, 249–254 (2004).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Acknowledgements
S.K.'s group research is supported by Agence Nationale de la Recherche EpiSperm and Institut National du Cancer funds. E.M. was supported by a three-year grant from the French Ministry of Research and an Association pour la Recherche sur le Cancer fellowship for her fourth-year PhD.
Author information
Authors and Affiliations
Contributions
Y.Z. and S.K. designed the experiments and wrote the paper. L.D. designed the experiments; synthesized chemicals; performed western blotting, immunoprecipitation, MS quantification and analysis; wrote the paper; and participated in other experiments. C.P. performed in vitro enzymatic assays. Y.C. and Z.D. performed MS analysis. E.M., Z.L., A.D., T.B., S.R., F.J., H.I. and B.R. performed immunofluorescence experiments, IHC and ChIP-Seq. B.R.S. and C.D.A. performed the Tetrahymena experiments.
Corresponding author
Ethics declarations
Competing interests
Y.Z. is a shareholder and a member of the scientific advisory board of PTM BioLabs, Co., Ltd. (Chicago, IL).
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–9, Supplementary Tables 1–5 and Supplementary Note. (PDF 3783 kb)
Rights and permissions
About this article
Cite this article
Dai, L., Peng, C., Montellier, E. et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol 10, 365–370 (2014). https://doi.org/10.1038/nchembio.1497
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1497
This article is cited by
-
Post-translational protein lactylation modification in health and diseases: a double-edged sword
Journal of Translational Medicine (2024)
-
Comparative study of 1H-NMR metabolomic profile of canine synovial fluid in patients affected by four progressive stages of spontaneous osteoarthritis
Scientific Reports (2024)
-
Proteome-Wide Analysis of Lysine 2-Hydroxyisobutyrylation in Aspergillus fumigatus
Current Microbiology (2024)
-
DeepKPred: Prediction and Functional Analysis of Lysine 2-Hydroxyisobutyrylation Sites Based on Deep Learning
Annals of Data Science (2024)
-
Proteomic analysis of lysine 2-hydroxyisobutyryl in SLE reveals protein modification alteration in complement and coagulation cascades and platelet activation Pathways
BMC Medical Genomics (2023)