Abstract
We genotyped individuals with primary biliary cirrhosis and unaffected controls for suggestive risk loci (genome-wide association P < 1 × 10−4) identified in a previous genome-wide association study. Combined analysis of the genome-wide association and replication datasets identified IRF5-TNPO3 (combined P = 8.66 × 10−13), 17q12-21 (combined P = 3.50 × 10−13) and MMEL1 (combined P = 3.15 × 10−8) as new primary biliary cirrhosis susceptibility loci. Fine-mapping studies showed that a single variant accounts for the IRF5-TNPO3 association. As these loci are implicated in other autoimmune conditions, these findings confirm genetic overlap among such diseases.
Similar content being viewed by others
Main
Primary biliary cirrhosis (PBC) is the most common autoimmune liver disease, characterized by chronic nonsuppurative destructive cholangitis, circulating anti-mitochondrial antibodies and the frequent presence of other autoimmune disorders in affected individuals and their family members1. We previously reported a genome-wide association study (GWAS) for PBC that identified three susceptibility loci, HLA, IL12A and IL12RB2, as being strongly associated with PBC, as well as a number of other loci showing suggestive association with this disease2. To replicate these findings and evaluate relevance of these latter loci to PBC susceptibility, we have now tested an additional independent cohort of 857 individuals with PBC (cases) of European descent and 3,198 controls of European descent (including 1,743 historic control subjects from the New York Cancer Project3) for PBC associations with 36 SNPs across 24 loci (Supplementary Fig. 1); for each locus, association signals at P < 1 × 10−4 were detected for more than one SNP found in the initial GWAS, with the exception of IRF5-TNPO3, a well-recognized autoimmune disease risk locus. The combination of these replication results and our prior genome-wide association data yielded a genetic dataset derived from 1,351 PBC cases and 4,700 controls (Supplementary Methods).
Fifteen SNPs replicated (P < 1.39 × 10−3) after Bonferroni adjustment (Table 1 and Supplementary Table 1). In addition to HLA, IL12A and IL12RB2, genes at the IRF5-TNPO3 locus, at chromosome 17q12-21 (containing IKZF3, ZPBP2, GSDMB and ORMDL3) and at MMEL1 loci showed significant association with PBC in both the replication analysis and in an analysis combining the replication and initial GWAS datasets (Fig. 1 and Supplementary Fig. 2).
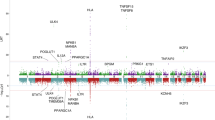
(a–c) Strength of the associations and recombination rates estimated from HapMap data for genotyped SNPs are shown for the (a) IRF5-TNPO3, (b) 17q12-21 and (c) MMEL1 loci. Both genome-wide association (circles) and combined fine-mapping (diamonds) data are shown where available. The extent of linkage disequilibrium with the most significant polymorphisms are indicated by the size of each data point; larger data points indicate stronger linkage disequilibrium. Gene positions for each gene region are indicated by the arrows, with the arrow direction representing the orientation of translation. Linkage disequilibrium was calculated using observed data in PLINK.
Among the PBC loci confirmed by this analysis, the locus at IRF5-TNPO3 (encoding interferon regulatory factor 5 and transportin 3) is of interest because of the integral immunoregulatory roles for IRF5, the prior association of this locus with systemic lupus erythematosus (SLE)4,5, systemic sclerosis6 and Sjögrens syndrome7, and the strong effect of the disease-associated SNP at this locus on disease risk (rs10488631) in the replication (P = 1.13 × 10−8, odds ratio (OR) = 1.58) and combined datasets (P = 8.66 × 10−13, OR = 1.57). Association of this locus was thus explored, initially by resequencing the IRF5 locus intronic and exonic regions and the 5′ and 3′ flanking regions of IRF5 in genomic pools of 100 subjects so as to delineate the genetic variation across the locus. This analysis confirmed prior reports of 41 polymorphisms across this region, but it failed to identify any new variants. Among these 41 SNPs, two (rs3807135 and rs3834330) failed repeatedly in genotyping assays and four (rs10275092, rs12537192, rs1727172 and rs754280) showed negligible polymorphism. Thus, for fine-mapping studies, 1,330 cases and 1,833 controls were genotyped for 35 SNPs across this region. Among the associations identified (Supplementary Table 2), the strongest signals were from the rs12539741 and rs2070197 alleles. These variants map to just 3′ of the IRF5 coding region and are in tight linkage disequilibrium with one another (r2 > 0.95), and their associations with PBC reach fine-mapping P values of 1.65 × 10−10 (OR = 1.63) and 3.74 × 10−10 (OR = 1.62), respectively. Conditional logistic regression analysis did not demonstrate any evidence for multiple genetic effects at the IRF5-TNPO3 locus, with the association at this locus with PBC appearing to be accounted for by a single variant, either rs12539741 or rs2070197 (Supplementary Table 3). Although unidentified variants outside the sequenced region may also be relevant to disease association or may even be disease causal, the latter two SNPs are among the SNPs at this locus that are highly associated with SLE, and both are correlated with changes in IRF5 expression in transformed B cells4. By contrast, an insertion-deletion polymorphism 64 bp upstream of IRF5 exon 1a, representing a putative disease-causal variant for SLE, showed modest association with PBC (fine-mapping P = 1.00 × 10−3). No associations were detected between PBC and another SLE-associated SNP, rs2004640, or a 3′ UTR variant, rs10954213, that has previously been identified as a predictor of IRF5 expression level4.
A second region of interest is the 17q12-21 locus, as all eight of the tested SNPs across this region achieved significance in the replication analysis (P values between 1.78 × 10−9 and 1.88 × 10−5). This chromosomal region has also been associated with asthma8, Crohn's disease9 and type 1 diabetes10 and contains four genes, ZPBP2 (encoding zona pellucida–binding protein 2), IKZF3 (encoding IKAROS family zinc finger 3 protein, involved in leukocyte development and IgE production), GSDMB (encoding gasdermin-B, involved in epithelial barrier function) and ORMDL3 (encoding ORM1-like protein 3). All eight of the SNPs tested here were in linkage disequilibrium (pairwise r2 values ranged from 0.66 to 0.96), but the strongest association signal came from a ZPBP2 SNP, rs11557467 (replication P = 1.78 × 10−9, OR = 0.71; combined P = 3.50 × 10−13, OR = 0.72). Although further studies are needed to pinpoint the relevant disease-causative allele(s), results of additive conditional logistic regression analysis suggest that this SNP fully accounts for the association signal across this region (Supplementary Table 4).
The replication and combined association data also identified a strong association of PBC with two SNPs (rs3890745 and rs3748816) at the MMEL1 (encoding membrane metallo-endopeptidase–like 1) locus on chromosome 1p36 (replication P = 3.31 × 10−6, OR = 1.31, combined P = 2.28 × 10−9, OR = 1.32 and replication P = 8.14 × 10−5, OR = 1.31, combined P = 3.15 × 10−8, OR = 1.33, respectively) (Table 1 and Supplementary Table 1). These SNPs are in linkage disequilibrium with one another (r2 > 0.88) and one of them (rs3748816) is a nonsynonymous SNP in exon 16 that encodes a potentially functional methionine-to-threonine substitution, whereas the other (rs3890745) maps within intron 2 of MMEL1 and has been associated with risk for rheumatoid arthritis and for celiac disease11,12.
A suggestive association signal (combined P = 9.12 × 10−7, OR = 1.27) was observed in the combined case-control cohort for rs3745516, an intronic SNP in SPIB, which is a gene encoding the Spi-B transcription factor. This SNP did not achieve significance in the replication cohort after Bonferroni correction (P = 6.21 × 10−3, OR = 1.18), but the strength of association in the combined analysis as well as the role of Spi-B in dendritic cell development13 and B-cell receptor signaling are in keeping with potential relevance of this locus to PBC pathogenesis.
Anti-mitochondrial antibodies (AMA) are found in most individuals with PBC, but no correlation of specific PBC genotypes with AMA status was observed in our prior2 or current association studies (data not shown). PBC is also associated with specific nuclear antibodies, with some 20% of individuals with PBC manifesting glycoprotein-210 antibodies (anti-gp210) directed against the human nuclear pore complex and/or sp100 antibodies that recognize a 53-kDa nuclear antigen14. Evaluation of genotype status in the subset of 462 cases typed for nuclear antibodies revealed a strong association of the HLA locus (rs9277535 at HLA-DPB1) with disease (P = 4.25 × 10−8, OR = 2.25) in anti-sp100–positive individuals (Supplementary Table 5a) in contrast to anti-sp100–negative individuals (P = 3.52 × 10−4, OR = 1.36). A further analysis incorporating GWAS data available for 412 of the individuals with known anti-sp100 status also revealed that among 13 MHC-region SNPs tested, three SNPs at the HLA-DPB1 locus were the most strongly associated with disease in anti-sp100–positive individuals but not in anti-sp100–negative individuals (Supplementary Table 5b). In particular, for these SNPs, the OR associations with PBC risk were much higher in anti-sp100–positive cases compared to anti-sp100–negative cases. By contrast, no genetic distinctions were apparent for the anti-gp210–positive subgroup (Supplementary Table 6). These findings need to be interpreted cautiously in view of the relatively small sample size used here, but they suggest a relevance of the HLA locus to anti-sp100 status, which parallels previously reported correlations of anti-CCP and HLA-DRB1 status in rheumatoid arthritis15.
In conclusion, we identify IRF5-TNPO3, 17q12-21 and MMEL1 as three new risk loci for PBC. Our data also suggest some genetic substructure may exist in PBC in relation to anti-sp100 status. Notably, all the PBC risk loci replicated or identified here have been implicated in other autoimmune diseases. Our data provide further evidence for the existence of shared autoimmunity susceptibility loci that contribute to the frequent appearance of additional autoimmune diseases in individuals with PBC and their families.
References
Lindor, K.D. et al. Hepatology 50, 291–308 (2009).
Hirschfield, G.M. et al. N. Engl. J. Med. 360, 2544–2555 (2009).
Mitchell, M.K., Gregersen, P.K., Johnson, S., Parsons, R. & Vlahov, D. J. Urban Health 81, 301–310 (2004).
Graham, R.R. et al. Proc. Natl. Acad. Sci. USA 104, 6758–6763 (2007).
Sigurdsson, S. et al. Hum. Mol. Genet. 17, 872–881 (2008).
Dieudé, P. et al. Arthritis Rheum. 60, 225–233 (2009).
Miceli-Richard, C. et al. Arthritis Rheum. 60, 1991–1997 (2009).
Bouzigon, E. et al. N. Engl. J. Med. 359, 1985–1994 (2008).
Barrett, J.C. et al. Nat. Genet. 40, 955–962 (2008).
Barrett, J.C. et al. Nat. Genet. 41, 703–707 (2009).
Raychaudhuri, S. et al. Nat. Genet. 40, 1216–1223 (2008).
Coenen, M.J. et al. Hum. Mol. Genet. 18, 4195–4203 (2009).
Schotte, R., Nagasawa, M., Weijer, K., Spits, H. & Blom, B. J. Exp. Med. 200, 1503–1509 (2004).
Muratori, L., Granito, A., Muratori, P., Pappas, G. & Bianchi, F.B. Clin. Liver Dis. vii. 12, 261–276 (2008).
Snir, O. et al. Ann. Rheum. Dis. 68, 736–743 (2009).
Acknowledgements
This research was supported by grants from the Canadian Institutes for Health Research (MOP 74621), the Ontario Research Fund (RE01-061), the Canadian Primary Biliary Cirrhosis Society, the Canadian Foundation for Innovation, the Ben and Hilda Katz Charitable Foundation, the US National Institutes of Health (K23 DK68290, RO3 DK78527 and RO1 DK80670), the American Gastroenterological Association and the A.J. and Sigismunda Palumbo Charitable Trust. K.A.S. is a recipient of a Canada Research Chair award. We thank all the study subjects and supporting physicians as well as the technical staff of the University Health Network Analytical Genetics Technology Centre for their assistance in this research.
Author information
Authors and Affiliations
Contributions
G.M.H., X.L., C.I.A. and K.A.S. designed the study. X.L., Y.H., I.P.G., Y.L., C.X., Y.L., W.C., J.Z. and C.I.A. performed the genotyping and data analysis. G.M.H., B.D.J., C.C., A.L.M., P.M., R.P.M., J.A.O., V.A.L., D.S., C.V., C.L., P.K.G., E.J.H., K.N.L. and K.A.S. developed the clinical network and sample collection and processing framework required for case and control accrual. G.M.H., C.I.A. and K.A.S. wrote the manuscript. G.M.H., X.L., C.I.A. and K.A.S. vouch for the data and its analysis. G.M.H., C.I.A. and K.A.S. (the senior and corresponding author) decided in agreement with all other authors to publish this paper.
Corresponding author
Ethics declarations
Competing interests
G.M.H. reports receiving lecture and consultancy fees from Axcan Pharma and A.L.M. received grant support from Novartis and Schering-Plough. J.H. reports receiving lecture fees and grant support from Axcan Pharma.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2, Supplementary Tables 1–6 and Supplementary Methods (PDF 1209 kb)
Rights and permissions
About this article
Cite this article
Hirschfield, G., Liu, X., Han, Y. et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet 42, 655–657 (2010). https://doi.org/10.1038/ng.631
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.631
This article is cited by
-
A regulatory variant at 19p13.3 is associated with primary biliary cholangitis risk and ARID3A expression
Nature Communications (2023)
-
The latest research trends in primary biliary cholangitis: a bibliometric analysis
Clinical and Experimental Medicine (2022)
-
Environmental factors, medical and family history, and comorbidities associated with primary biliary cholangitis in Japan: a multicenter case–control study
Journal of Gastroenterology (2022)
-
rs1944919 on chromosome 11q23.1 and its effector genes COLCA1/COLCA2 confer susceptibility to primary biliary cholangitis
Scientific Reports (2021)