Abstract
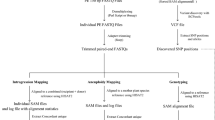
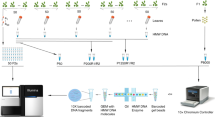
We describe 2b-RAD, a streamlined restriction site–associated DNA (RAD) genotyping method based on sequencing the uniform fragments produced by type IIB restriction endonucleases. Well-studied accessions of Arabidopsis thaliana were genotyped to validate the method's accuracy and to demonstrate fine-tuning of marker density as needed. The simplicity of the 2b-RAD protocol makes it particularly suitable for high-throughput genotyping as required for linkage mapping and profiling genetic variation in natural populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gray, I.C., Campbell, D.A. & Spurr, N.K. Hum. Mol. Genet. 9, 2403–2408 (2000).
Morin, P.A., Luikart, G., Wayne, R.K. & the SNP Workshop Group. Trends Ecol. Evol. 19, 208–216 (2004).
Clark, A.G., Hubisz, M.J., Bustamante, C.D., Williamson, S.H. & Nielsen, R. Genome Res. 15, 1496–1502 (2005).
Maher, B. Nature 456, 18–21 (2008).
Manolio, T.A. et al. Nature 461, 747–753 (2009).
Davey, J.W. et al. Nat. Rev. Genet. 12, 499–510 (2011).
Baird, N.A. et al. PLoS ONE 3, e3376 (2008).
Chutimanitsakun, Y. et al. BMC Genomics 12, 4 (2011).
Hohenlohe, P.A. et al. PLoS Genet. 6, e1000862 (2010).
Emerson, K.J. et al. Proc. Natl. Acad. Sci. USA 107, 16196–16200 (2010).
Catchen, J., Amores, A., Hohenlohe, P., Cresko, W. & Postlethwait, J. G3 (Bethesda) 1, 171–182 (2011).
Van Tassell, C.P. et al. Nat. Methods 5, 247–252 (2008).
van Orsouw, N.J. et al. PLoS ONE 2, e1172 (2007).
Elshire, R.J. et al. PLoS ONE 6, e19379 (2011).
Clark, R.M. et al. Science 317, 338–342 (2007).
Mckay, J.K., Richards, J.H. & Mitchell-Olds, T. Mol. Ecol. 12, 1137–1151 (2003).
Clauss, M.J., Cobban, H. & Mitchell-Olds, T. Mol. Ecol. 11, 591–601 (2002).
Rumble, S.M. et al. PLoS Comput. Biol. 5, e1000386 (2009).
Li, W. & Godzik, A. Bioinformatics 22, 1658–1659 (2006).
Li, R. et al. Bioinformatics 25, 1966–1967 (2009).
Li, R. et al. Genome Res. 19, 1124–1132 (2009).
Acknowledgements
This work was supported by the National Science Foundation grants DEB-1054766 to M.V.M. and DEB-1022196 to J.K.M., and by grants from the National Natural Science Foundation of China (31130054) and National High Technology Research and Development Program of China (2012AA10A405) to S.W. We are grateful to T.E. Juenger and D.L. Des Marais (University of Texas at Austin) for growing Arabidopsis samples.
Author information
Authors and Affiliations
Contributions
S.W. and M.V.M. conceived and designed the study. S.W. developed the original protocol for 2b-RAD library preparation, and E.M. developed modifications for RTR. S.W. and E.M. prepared 2b-RAD libraries for SOLiD and Illumina sequencing. E.M. created bioinformatics scripts and conducted sequence analysis and genotype validation. J.K.M. provided Arabidopsis samples and contributed resequencing data for bioinformatic validation. S.W., E.M., J.K.M. and M.V.M. wrote the paper. M.V.M. supervised the whole study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 , Supplementary Tables 1–11 and Supplementary Protocol (PDF 365 kb)
Supplementary Software
Perl scripts for 2b-RAD analysis. (ZIP 12 kb)
Rights and permissions
About this article
Cite this article
Wang, S., Meyer, E., McKay, J. et al. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat Methods 9, 808–810 (2012). https://doi.org/10.1038/nmeth.2023
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2023
This article is cited by
-
2bRAD reveals fine-scale genetic structuring among populations within the Mediterranean zoanthid Parazoanthus axinellae (Schmidt, 1862)
Coral Reefs (2024)
-
Genome-wide SNP markers provided insights into the reproductive strategy and genetic diversity of the green tide causative species Ulva prolifera in China
Journal of Oceanology and Limnology (2024)
-
Global patterns of genomic and phenotypic variation in the invasive harlequin ladybird
BMC Biology (2023)
-
Oral microbiota disorder in GC patients revealed by 2b-RAD-M
Journal of Translational Medicine (2023)
-
Gastrointestinal symptoms of long COVID-19 related to the ectopic colonization of specific bacteria that move between the upper and lower alimentary tract and alterations in serum metabolites
BMC Medicine (2023)