Abstract
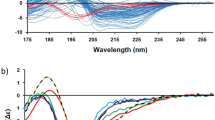
Circular dichroism (CD) is an excellent tool for rapid determination of the secondary structure and folding properties of proteins that have been obtained using recombinant techniques or purified from tissues. The most widely used applications of protein CD are to determine whether an expressed, purified protein is folded, or if a mutation affects its conformation or stability. In addition, it can be used to study protein interactions. This protocol details the basic steps of obtaining and interpreting CD data, and methods for analyzing spectra to estimate the secondary structural composition of proteins. CD has the advantage that measurements may be made on multiple samples containing ≤20 μg of proteins in physiological buffers in a few hours. However, it does not give the residue-specific information that can be obtained by x-ray crystallography or NMR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Velluz, L., Legrand, M. & Grosjean, M. Optical Circular Dichroism: Principles, Measurements, and Applications (Verlag Chemie Academic Press Inc., New York and London, 1965).
Beychok, S. Circular dichroism of biological macromolecules. Science 154, 1288–1299 (1966).
Sreerama, N. & Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383, 318–351 (2004).
Holzwarth, G. & Doty, P. The ultraviolet circular dichroism of polypeptides. J. Am. Chem. Soc. 87, 218–228 (1965).
Greenfield, N. & Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8, 4108–4116 (1969).
Venyaminov, S., Baikalov, I.A., Shen, Z.M., Wu, C.S. & Yang, J.T. Circular dichroic analysis of denatured proteins: inclusion of denatured proteins in the reference set. Anal. Biochem. 214, 17–24 (1993).
Bovey, F.A. & Hood, F.P. Circular dichroism spectrum of poly-L-proline. Biopolymers 5, 325–326 (1967).
Tiffany, M.L. & Krimm, S. Effect of temperature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers 11, 2309–2316 (1972).
Woody, R.W. Circular dichroism and conformation of unordered polypeptides. Adv. Biophys. Chem. 2, 31–79 (1992).
Rucker, A.L. & Creamer, T.P. Polyproline II helical structure in protein unfolded states: lysine peptides revisited. Protein Sci. 11, 980–985 (2002).
Nafie, L.A. Infrared and Raman vibrational optical activity: theoretical and experimental aspects. Annu. Rev. Phys. Chem. 48, 357–386 (1997).
Jackson, M. & Mantsch, H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 30, 95–120 (1995).
Cooper, E.A. & Knutson, K. Fourier transform infrared spectroscopy investigations of protein structure. Pharm. Biotechnol. 7, 101–143 (1995).
Pelton, J.T. & McLean, L.R. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 277, 167–176 (2000).
Lowry, O.H., Rosebrough, N.J., Farr, A.L. & Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254 (1976).
Hennessey, J.P. Jr. & Johnson, W.C. Jr. Information content in the circular dichroism of proteins. Biochemistry 20, 1085–1094 (1981).
Lang, C.A. Simple microdetermination of Kjeldahl nitrogen in biological material. Anal. Chem. 30, 1692–1694 (1958).
Jaenicke, L. A rapid micromethod for the determination of nitrogen and phosphate in biological material. Anal. Biochem. 61, 623–627 (1974).
Brahms, S. & Brahms, J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J. Mol. Biol. 138, 149–178 (1980).
Reed, J. & Reed, T.A. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal. Biochem. 254, 36–40 (1997).
Saxena, V.P. & Wetlaufer, D.B. A new basis for interpreting the circular dichroic spectra of proteins. Proc. Natl Acad. Sci. USA 68, 969–972 (1971).
Chen, Y.H., Yang, J.T. & Chau, K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13, 3350–3359 (1974).
Chang, C.T., Wu, C.S. & Yang, J.T. Circular dichroic analysis of protein conformation: inclusion of the β-turns. Anal. Biochem. 91, 13–31 (1978).
Yang, J.T., Wu, C.S. & Martinez, H.M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 130, 208–269 (1986).
Greenfield, N.J. Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal. Biochem. 235, 1–10 (1996).
Provencher, S.W. & Glöckner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 (1981).
Manavalan, P. & Johnson, W.C. Jr. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167, 76–85 (1987).
Toumadje, A., Alcorn, S.W. & Johnson, W.C. Jr. Extending CD spectra of proteins to 168 nm improves the analysis for secondary structures. Anal. Biochem. 200, 321–331 (1992).
Toumadje, A. & Johnson, W.C. Jr. Effects of relative band intensity on prediction of protein secondary structure from CD. Anal. Biochem. 211, 258–260 (1993).
Johnson, W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35, 307–312 (1999).
Sreerama, N. & Woody, R.W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 (1993).
Sreerama, N. & Woody, R.W. Protein secondary structure from circular dichroism spectroscopy. Combining variable selection principle and cluster analysis with neural network, ridge regression and self-consistent methods. J. Mol. Biol. 242, 497–507 (1994).
Sreerama, N., Venyaminov, S.Y. & Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8, 370–380 (1999).
Sreerama, N., Venyaminov, S.Y. & Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 287, 243–251 (2000).
Sreerama, N. & Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000).
Böhm, G., Muhr, R. & Jaenicke, R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5, 191–195 (1992).
Andrade, M.A., Chacón, P., Merelo, J.J. & Morán, F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6, 383–390 (1993).
Unneberg, P., Merelo, J.J., Chacón, P. & Morán, F. SOMCD: method for evaluating protein secondary structure from UV circular dichroism spectra. Proteins 42, 460–470 (2001).
Perczel, A., Park, K. & Fasman, G.D. Analysis of the circular dichroism spectrum of proteins using the convex constraint algorithm: a practical guide. Anal. Biochem. 203, 83–93 (1992).
Chen, Y.H. & Yang, J.T. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem. Biophys. Res. Commun. 44, 1285–1291 (1971).
Chen, Y.H., Yang, J.T. & Martinez, H.M. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11, 4120–4131 (1972).
Sreerama, N. & Woody, R.W. Poly(pro)II helices in globular proteins: identification and circular dichroic analysis. Biochemistry 33, 10022–10025 (1994).
Perczel, A., Hollósi, M., Tusnády, G. & Fasman, G.D. Convex constraint analysis: a natural deconvolution of circular dichroism curves of proteins. Protein Eng. 4, 669–679 (1991).
Greenfield, N.J. & Hitchcock-DeGregori, S.E. Conformational intermediates in the folding of a coiled-coil model peptide of the N-terminus of tropomyosin and α-tropomyosin. Protein Sci. 2, 1263–1273 (1993).
Safar, J., Roller, P.P., Gajdusek, D.C. & Gibbs, C.J. Jr. Thermal stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 2, 2206–2216 (1993).
Greenfield, N.J. Using circular dichroism, collected as a function of temperature, to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. doi: 10.1038/nprot.2006.204 (2006).
Adler, A.J., Greenfield, N.J. & Fasman, G.D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 27, 675–735 (1973).
Rosenkranz, H. Circular dichroism of helical and nonhelical proteins. A review of the limits of the methods for calculating secondary structure. Z. Klin. Chem. Klin. Biochem. 12, 222 (1974).
Johnson, W.C. Jr. Protein secondary structure and circular dichroism: a practical guide. Proteins 7, 205–214 (1990).
Woody, R.W. Circular dichroism. Methods Enzymol. 246, 34–71 (1995).
Kelly, S.M., Jess, T.J. & Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 (2005).
Miles, A.J. & Wallace, B.A. Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 35, 39–51 (2006).
Venyaminov, S.Y. & Yang,, J.T. in Circular Dichroism and the Conformational Analysis of Biomolecules (ed. Fasman, G.D.) (Plenum Press, New York and London, 1996).
Greenfield, N.J. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat. Protoc. doi: 10.1038/nprot.2006.229 (2006).
Greenfield, N.J. The use of circular dichroism to study the kinetics of protein folding and unfolding. Nat. Protoc. doi: 10.1038/nprot.2006.244 (2006).
Miles, A.J., Whitmore, L. & Wallace, B.A. Spectral magnitude effects on the analyses of secondary structure from circular dichroism spectroscopic data. Protein Sci. 14, 368–374 (2005).
Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 (1967).
Mihalyi, E. Numerical values of the absorbances of the aromatic amino acids in acid, neutral, and alkaline solutions. J. Chem. Eng. Data 13, 179–182 (1968).
Goa, J. A micro biuret method for protein determination. Scand. J. Clin. Lab. Invest. 5, 218–222 (1953).
Konno, T. Conformational diversity of acid-denatured cytochrome c studied by a matrix analysis of far-UV CD spectra. Protein Sci. 7, 975–982 (1998).
Melberg, S.G. & Johnson, W.C. Jr. Changes in secondary structure follow the dissociation of human insulin hexamers: a circular dichroism study. Proteins 8, 280–286 (1990).
Savitsky, A. & Golay, M.J.E. Smoothing and dIfferentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964).
Bentz, H., Bächinger, H.P., Glanville, R. & Kühn, K. Physical evidence for the assembly of A and B chains of human placental collagen in a single triple helix. Eur. J. Biochem. 92, 563–567 (1978).
Steiner, R.F. Structural transitions of lysozyme. Biochim. Biophys. Acta 79, 51–63 (1964).
Acknowledgements
I thank M.A. Andrade, W.C. Johnson, Jr., N. Sreerama, R.W. Woody, S. Venyaminov, and especially the late Gerald D. Fasman for supplying versions of their software for analyzing CD data for use, analysis and distribution. The research was supported by a National Institutes of Health grant, GM-36326 to N.J.G. and to Sarah E. Hitchcock-DeGregori and by the Circular Dichroism Facility at Robert Wood Johnson Medical School (UMDNJ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Greenfield, N. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc 1, 2876–2890 (2006). https://doi.org/10.1038/nprot.2006.202
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.202
This article is cited by
-
Engineered ice-binding protein (FfIBP) shows increased stability and resistance to thermal and chemical denaturation compared to the wildtype
Scientific Reports (2024)
-
Perfect circular polarization of elastic waves in solid media
Nature Communications (2024)
-
ATP-dependent conformational dynamics in a photoactivated adenylate cyclase revealed by fluorescence spectroscopy and small-angle X-ray scattering
Communications Biology (2024)
-
Revealing the mechanistic interactions of profenofos and captan pesticides with serum protein via biophysical and computational investigations
Scientific Reports (2024)
-
Inter-coat protein loading of active ingredients into Tobacco mild green mosaic virus through partial dissociation and reassembly of the virion
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.