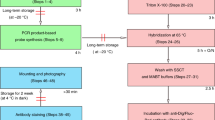
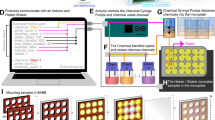
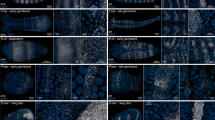
Abstract
We describe a whole-mount RNA in situ hybridization (ISH) method optimized for detection of the cellular and subcellular distributions of specific mRNA within Drosophila testes and male genital tract. Digoxygenin (dig)-labeled antisense RNA probes are in vitro transcribed from a template synthesized by (RT)-PCR; the probe length is reduced by hydrolysis. Testes and male genital tracts are dissected from adult flies, fixed and processed for hybridization. Both probe and fixed testes can be stored before use. Extensive post-hybridization washing reduces the background. Detection is through alkaline phosphatase-conjugated anti-dig antibodies followed by a color reaction. This protocol is suitable for low-medium throughput applications with parallel processing of 2–48 samples, and takes 4–5 d to complete. We have used this protocol, which is similar to other RNA ISH protocols, but optimized for whole-mount Drosophila testes, to document the expression of about 1,000 genes in Drosophila melanogaster male genital tract.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Fuller, M.T. Spermatogenesis. In The Development of Drosophila. (eds. Bate, M. & Martinez-Arias, A.) 71–147 (Cold Spring Harbor Press, Cold Spring Harbor, New York, 1993).
Barreau, C., Benson, E., Gudmannsdottir, E., Newton, F. & White-Cooper, H. Post-meiotic transcription in Drosophila testes. Development 135, 1897–1902 (2008).
Olivieri, G. & Olivieri, A. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster . Mutat. Res. 2, 366–380 (1965).
Schäfer, M., Nayernia, K., Engel, W. & Schäfer, U. Translational control in spermatogenesis. Dev. Biol. 172, 344–352 (1995).
Hales, K.G. & Fuller, M.T. Developmentally regulated mitochondrial fusion mediated by a conserved novel predicted GTPase. Cell 90, 121–129 (1997).
Vardanyan, A. et al. Dumpy-30 family members as determinants of male fertility and interaction partners of metal-responsive transcription factor 1 (MTF-1) in Drosophila . BMC Dev. Biol. 8, 68 (2008).
Kuhn, R., Schafer, U. & Schafer, M. Cis-acting regions sufficient for spermatocyte-specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9 . EMBO J. 7, 447–454 (1988).
White-Cooper, H., Schafer, M.A., Alphey, L.S. & Fuller, M.T. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila . Development 125, 125–134 (1998).
Chintapalli, V., Wang, J. & Dow, J. Using FlyAtlas to identify better Drosophila models of human disease. Nat. Genet. 39, 715–720 (2007).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback . Chromosoma 98, 81–85 (1989).
Lehmann, R. & Tautz, D. In situ hybridization to RNA. Methods Cell Biol. 44, 575–598 (1994).
Roche. Nonradioactive In Situ Hybridization Application Manual (Roche Diagnostics GmbH, Penzberg, Germany, 2002).
Alphey, L. et al. twine, a cdc25 homologue that functions in the male and female germlines of Drosophila . Cell 69, 977–988 (1992).
Wang, F. et al. Laminin in hte male germ cells of Drosophila . J. Cell Biol. 119, 977–988 (1992).
Lecuyer, E. et al. Global analysis of mRNA localisation reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007).
Robida, M.D. & Singh, R. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J. 22, 2924–2933 (2003).
Harhangi, H.R. et al. RADHA—a new male germ line-specific chromosomal protein of Drosophila . Chromosoma 108, 235–242 (1999).
Rothwell, W.F. & Sullivan, W. Fluorescent analysis of Drosophila embryos. In Drosophila Protocols (eds. Sullivan, W., Ashburner, M. & Hawley, R.S.) 141–157 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2000).
White-Cooper, H. Spermatogenesis: Analysis of meiosis and morphogenesis. In Drosophila Cytogenetics Protocols. (ed. Henderson, D.) 45–75 (Humana Press, Totowa, New Jersey, 2004).
Weiszmann, R., Hammonds, A.S. & Celniker, S.E. Determination of gene expression patterns using high-throughput RNA in situ hybridisation to whole-mount Drosophila embryos. Nat. Protoc. 4, 605–618 (2009).
Acknowledgements
We thank the succession of project students, technicians, PhD students and post-docs who have over the years carried out RNA in situ hybridization in the lab. Between all of us we have probably made every mistake possible in this protocol, thus identifying the important steps. We are very grateful for support from the UK Biotechnology and Biological Sciences Research Council for our testis RNA in situ hybridization projects (Grants BB/C503903/1 and BB_D009324_1). H.W.-C. is a Royal Society University Research Fellow.
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper. H.W.-C. adapted and optimized the non-radioactive ISH methods from Drosophila ovaries and testes to work reliably and with minimal background in Drosophila testes and contributed extensively to the paper writing. E.B. previously performed many experiments with the method and contributed to making it a robust assay, particularly by teaching others. She acted as the 'experienced operator' and gave feedback/technical tips on the protocol/paper. C.M. acted as the 'inexperienced operator,' contributed extensively to the writing of the paper and ran many of the experiments used in the figures, all of which are novel and have not been previously published.
Corresponding author
Rights and permissions
About this article
Cite this article
Morris, C., Benson, E. & White-Cooper, H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protoc 4, 1807–1819 (2009). https://doi.org/10.1038/nprot.2009.192
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.192
This article is cited by
-
Cellular metabolic reprogramming controls sugar appetite in Drosophila
Nature Metabolism (2020)
-
Drosophila Pif1A is essential for spermatogenesis and is the homolog of human CCDC157, a gene associated with idiopathic NOA
Cell Death & Disease (2019)
-
‘Escaping’ the X chromosome leads to increased gene expression in the male germline of Drosophila melanogaster
Heredity (2014)
-
Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries
Nature Protocols (2013)
-
Dual fluorescence detection of protein and RNA in Drosophila tissues
Nature Protocols (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.