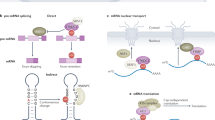
Key Points
-
RNA modifications may post-transcriptionally regulate RNA stability, localization, transport, splicing and translation.
-
METTL3 and METTL14 catalyse N6-methyladenosine (m6A) methylation of mRNA (and other types of nuclear RNA) both in vitro and in vivo. Wilms' tumour 1-associating protein (WTAP) is another crucial component of this methyltransferase complex.
-
α-ketoglutarate-dependent dioxygenases FTO and ALKBH5 are m6A demethylases of mRNA (and other types of nuclear RNA) that affect biological processes such as development, energy homeostasis and spermatogenesis.
-
Genome-wide mapping of m6A in mRNA reveals that m6A localizes around stop codons and at 3′ untranslated regions in mammals and yeast. The methylation is dynamic and seems to have regulatory roles.
-
The YTHDF domain family proteins preferentially bind to m6A in mRNA. The recognition of m6A in mRNA and other polyadenylated RNA by YTHDF2 reduces the half-lives of its substrate RNAs through processing-body-mediated degradation.
-
RNA methylation directly affects the cell circadian cycle, embryonic stem cell differentiation and yeast meiosis.
-
We propose that the reversible RNA methylation pathway has evolved to regulate processes that involve rapid expression changes of large groups of genes and proteins.
Abstract
Cellular RNAs carry diverse chemical modifications that used to be regarded as static and having minor roles in 'fine-tuning' structural and functional properties of RNAs. In this Review, we focus on reversible methylation through the most prevalent mammalian mRNA internal modification, N6-methyladenosine (m6A). Recent studies have discovered protein 'writers', 'erasers' and 'readers' of this RNA chemical mark, as well as its dynamic deposition on mRNA and other types of nuclear RNA. These findings strongly indicate dynamic regulatory roles that are analogous to the well-known reversible epigenetic modifications of DNA and histone proteins. This reversible RNA methylation adds a new dimension to the developing picture of post-transcriptional regulation of gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Suzuki, M. M. & Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nature Rev. Genet. 9, 465–476 (2008).
Kohli, R. M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Rev. Genet. 13, 484–492 (2012).
Branco, M. R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature Rev. Genet. 13, 7–13 (2012).
Bhutani, N., Burns, D. M. & Blau, H. M. DNA demethylation dynamics. Cell 146, 866–872 (2011).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Shi, Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nature Rev. Genet. 8, 829–833 (2007).
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev. Genet. 7, 715–727 (2006).
Bird, A. Molecular biology. Methylation talk between histones and DNA. Science. 294, 2113–2115 (2001).
He, C. Grand challenge commentary: RNA epigenetics? Nature Chem. Biol. 6, 863–865 (2010).
Grosjean, H. & Benne, R. Modification and Editing of RNA (American Society for Microbiology Press, 1998).
Grosjean, H. Fine-Tuning of RNA Functions by Modification and Editing (Springer-Verlag, 2005).
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways — 2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Motorin, Y. & Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631 (2011).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chem. Biol. 7, 885–887 (2011). This work describes a major breakthrough of discovering the first m6A RNA demethylase FTO, which highlights the possible biological function of m6A.
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). This study discovered the second mammalian m6A demethylase ALKBH5 that affects mouse spermatogenesis.
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012). References 17 and 18 revealed, for the first time, the transcriptome-wide distributions of m6A in mammalian genomes.
Wei, C. M., Gershowitz, A. & Moss, B. Methylated nucleotides block 5′ terminus of HeLa-cell messenger-RNA. Cell 4, 379–386 (1975).
Krug, R. M., Morgan, M. A. & Shatkin, A. J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 20, 45–53 (1976).
Rottman, F. M., Desrosiers, R. C. & Friderici, K. Nucleotide methylation patterns in eukaryotic mRNA. Prog. Nucleic Acid. Res. Mol. Biol. 19, 21–38 (1976).
Beemon, K. & Keith, J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 113, 165–179 (1977).
Schibler, U., Kelley, D. E. & Perry, R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 115, 695–714 (1977).
Wei, C. M. & Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Narayan, P. & Rottman, F. M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 242, 1159–1162 (1988).
Csepany, T., Lin, A., Baldick, C. J. Jr & Beemon, K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 265, 20117–20122 (1990).
Narayan, P., Ludwiczak, R. L., Goodwin, E. C. & Rottman, F. M. Context effects on N6-adenosine methylation sites in prolactin mRNA. Nucleic Acids Res. 22, 419–426 (1994).
Rottman, F., Shatkin, A. J. & Perry, R. P. Sequences containing methylated nucleotides at 5′ termini of messenger-RNAs — possible implications for processing. Cell 3, 197–199 (1974).
Bodi, Z., Button, J. D., Grierson, D. & Fray, R. G. Yeast targets for mRNA methylation. Nucleic Acids Res. 38, 5327–5335 (2010).
Keith, G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 77, 142–144 (1995).
Clancy, M. J., Shambaugh, M. E., Timpte, C. S. & Bokar, J. A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002).
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013). This study reveals the dynamics of transcriptome-wide m6A changes during yeast meiosis.
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Carroll, S. M., Narayan, P. & Rottman, F. M. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell. Biol. 10, 4456–4465 (1990).
Kierzek, E. & Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480 (2003).
Harcourt, E. M., Ehrenschwender, T., Batista, P. J., Chang, H. Y. & Kool, E. T. Identification of a selective polymerase enables detection of N6-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013).
Vilfan, I. D. et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J. Nanobiotechnol. 11, 8 (2013).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997). This pivotal study identifies METTL3 as a key SAM-binding subunit of the RNA methyltransferase complex.
Bokar, J. A. in Fine-Tuning of RNA Functions by Modification and Editing 141–177 (Springer-Verlag, 2005).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chem. Biol. 10, 93–95 (2014). This paper uncovers the core components of the m6A RNA methyltransferase complex and reveals an overall negative correlation between the levels of m6A mRNA methylation and gene expression.
Bujnicki, J. M., Feder, M., Radlinska, M. & Blumenthal, R. M. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 55, 431–444 (2002).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biol. 16, 191–198 (2014). This study discovered that the m6A modification on mRNA affects embryonic cell differentiation.
Alexandrov, A., Martzen, M. R. & Phizicky, E. M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8, 1253–1266 (2002).
Chujo, T. & Suzuki, T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 (2012).
Ozanick, S., Krecic, A., Andersland, J. & Anderson, J. T. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290 (2005).
Leulliot, N. et al. Structure of the yeast tRNA m7G methylation complex. Structure 16, 52–61 (2008).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008).
Agarwala, S. D., Blitzblau, H. G., Hochwagen, A. & Fink, G. R. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 8, e1002732 (2012).
Little, N. A., Hastie, N. D. & Davies, R. C. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum. Mol. Genet. 9, 2231–2239 (2000).
Ping, X. L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Horiuchi, K. et al. Identification of Wilms' Tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288, 33292–33302 (2013).
Bodi, Z. et al. Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3, 48 (2012).
Hongay, C. F. & Orr-Weaver, T. L. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl Acad. Sci. USA 108, 14855–14860 (2011).
Peters, T., Ausmeier, K. & Ruther, U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm. Genome 10, 983–986 (1999).
Dina, C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genet. 39, 724–726 (2007).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Scuteri, A. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007).
Gerken, T. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 (2007).
Fischer, J. et al. Inactivation of the Fto gene protects from obesity. Nature 458, 894–898 (2009).
Church, C. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nature Genet. 42, 1086–1092 (2010).
Boissel, S. et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 85, 106–111 (2009).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Jia, G. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319 (2008).
Hess, M. E. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nature Neurosci. 16, 1042–1048 (2013).
Gulati, P. et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl Acad. Sci. USA 110, 2557–2562 (2013).
Han, Z. et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464, 1205–1209 (2010).
Zheng, G. et al. Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases. RNA Biol. 10, 915–918 (2013).
Baltz, A. G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Fu, Y. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature Commun. 4, 1798 (2013).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nature Biotech. 29, 436–442 (2011).
Robbens, S. et al. The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J. Mol. Evol. 66, 80–84 (2008).
Iyer, L. M., Tahiliani, M., Rao, A. & Aravind, L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8, 1698–1710 (2009).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). This work presents the first m6A reader protein to be characterized, YTHDF2, and a main function of m6A: YTHDF2 mediates the m6A-dependent RNA decay by targeting RNA substrates to P-bodies.
Schoenberg, D. R. & Maquat, L. E. Regulation of cytoplasmic mRNA decay. Nature Rev. Genet. 13, 246–259 (2012).
Isken, O. & Maquat, L. E. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nature Rev. Genet. 9, 699–712 (2008).
Sheth, U. & Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 (2003).
Han, D. et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575 (2009).
Marzluff, W. F., Wagner, E. J. & Duronio, R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature Rev. Genet. 9, 843–854 (2008).
Dasgupta, T. & Ladd, A. N. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 3, 104–121 (2012).
Yang, F. & Schoenberg, D. R. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell 14, 435–445 (2004).
Ghosh, S. & Jacobson, A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip. Rev. RNA 1, 351–361 (2010).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Ameres, S. L. & Zamore, P. D. Diversifying microRNA sequence and function. Nature Rev. Mol. Cell Biol. 14, 475–488 (2013).
Harigaya, Y. et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50 (2006).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Kawai, T. & Akira, S. Toll-like receptor and RIG-I-like receptor signaling. Ann. NY Acad. Sci. 1143, 1–20 (2008).
Newby, M. I. & Greenbaum, N. L. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nature Struct. Biol. 9, 958–965 (2002).
Lebedeva, S. et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352 (2011).
Mukherjee, N. et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339 (2011).
Srikantan, S. & Gorospe, M. UneCLIPsing HuR nuclear function. Mol. Cell 43, 319–321 (2011).
Dormoy-Raclet, V. et al. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA. Nature Commun. 4, 2388 (2013).
Barnhart, M. D., Moon, S. L., Emch, A. W., Wilusz, C. J. & Wilusz, J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 5, 909–917 (2013).
Abdelmohsen, K. & Gorospe, M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 1, 214–229 (2010).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Chen, K. & Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nature Rev. Genet. 8, 93–103 (2007).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 9, 102–114 (2008).
Parker, R. & Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (2007).
Keene, J. D. RNA regulons: coordination of post-transcriptional events. Nature Rev. Genet. 8, 533–543 (2007).
Gallego, M. & Virshup, D. M. Post-translational modifications regulate the ticking of the circadian clock. Nature Rev. Mol. Cell Biol. 8, 139–148 (2007).
Eulalio, A., Behm-Ansmant, I. & Izaurralde, E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 8, 9–22 (2007).
Fustin, J. M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013). This study shows that the m6A modification affects the export of several mRNAs that are related to the circadian cycle.
Khan, Z. et al. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342, 1100–1104 (2013).
Wu, L. et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82 (2013).
Saletore, Y. et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012).
Karijolich, J. & Yu, Y. T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011).
Fernandez, I. S. et al. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500, 107–110 (2013).
Ge, J. & Yu, Y. T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 38, 210–218 (2013).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Hussain, S., Aleksic, J., Blanco, S., Dietmann, S. & Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14, 215 (2013).
Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Bykhovskaya, Y., Casas, K., Mengesha, E., Inbal, A. & Fischel-Ghodsian, N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 74, 1303–1308 (2004).
Patton, J. R., Bykhovskaya, Y., Mengesha, E., Bertolotto, C. & Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 280, 19823–19828 (2005).
Sahoo, T. et al. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nature Genet. 40, 719–721 (2008).
Sedgwick, B. Repairing DNA-methylation damage. Nature Rev. Mol. Cell Biol. 5, 148–157 (2004).
Mishina, Y., Duguid, E. M. & He, C. Direct reversal of DNA alkylation damage. Chem. Rev. 106, 215–232 (2006).
Fu, Y. et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed Engl. 49, 8885–8888 (2010).
van den Born, E. et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nature Commun. 2, 172 (2011).
Aik, W. et al. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. http://dx.doi.org/10.1093/nar/gku085 (2014).
Chen, W. et al. Crystal structure of the RNA demethylase ALKBH5 from zebrafish. FEBS Lett. 588, 892–898 (2014).
Acknowledgements
The authors apologize to colleagues whose work was not cited owing to space limitation. They thank T. Pan, X. Wang, Y. Yue and J. Liu for discussion. C.H. is supported by the US National Institutes of Health grants GM071440 and the EUREKA grant GM088599. This work was also supported partly by grants from the Israel Science Foundation, the Flight Attendant Medical Research Institute (FAMRI) and the Israeli Centers of Research Excellence. S.F. Reichard contributed to editing of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Epigenetic modifications
-
Reversible chemical modifications on DNA and histones that regulate gene expression independently of the genome sequences and that are heritable through cell division.
- 'Writers', 'erasers' and 'readers'
-
Enzymes or proteins that add, remove or preferentially bind to the chemical modifications at designated DNA or RNA nucleotides and amino acid residues of histones.
- Methyltransferase
-
An enzyme that transfers a methyl group to its substrate. Most methyltransferases use S-adenosyl-l-methionine (SAM) as the methyl donor.
- Two-dimensional thin layer chromatography
-
A technique to separate and identify nucleosides on cellulose plates according to their differential migration patterns in two different solvents. The nucleoside is typically radiolabelled for detection.
- High-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry
-
(HPLC–QqQ-MS/MS). A liquid chromatography method coupled with triple-quadrupole tandem mass spectrometry, which can quantitatively and simultaneously monitor multiple molecular species according to their fragmentation patterns.
- m6A RNA immunoprecipitation
-
An immunoprecipitation method to selectively enrich for N6-methyladenosine (m6A)-containing RNA using an m6A-targeted antibody.
- Nuclear speckles
-
Nuclear domains located in the interchromatin regions of the nucleoplasm and enriched with pre-mRNA processing factors.
- Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation
-
(PAR–CLIP). A biochemical method that takes advantage of incorporated photoreactive ribonucleoside analogues to identify the binding sites of RNA-binding proteins in cells.
- Yeast two-hybrid screens
-
A method in which one protein is fused to the GAL4 activation domain and the other to the GAL4 DNA-binding domain, and both fusion proteins are introduced into yeast. Expression of a GAL4-regulated reporter gene indicates that the two proteins physically interact.
- Demethylase
-
An enzyme that removes a methyl group from its substrate.
- Oxidative demethylation
-
A chemical reaction in which the C–H bond of a methyl group attached to a nitrogen or an oxygen atom is oxidized to –OH by demethylases, and the intermediate decomposes to release the methyl group as formaldehyde.
- Ribosome profiling
-
Qualitative and quantitative sequencing of the RNA attached to ribosomes as a signature of genes that are expressed.
- Processing bodies
-
(P-bodies). Distinct foci in the cytoplasm that are enriched with RNA degradation factors.
Rights and permissions
About this article
Cite this article
Fu, Y., Dominissini, D., Rechavi, G. et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 15, 293–306 (2014). https://doi.org/10.1038/nrg3724
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3724
This article is cited by
-
Circular RNA-circPan3 attenuates cardiac hypertrophy via miR-320-3p/HSP20 axis
Cellular & Molecular Biology Letters (2024)
-
N6-methyladenosine modification positively regulate Japanese encephalitis virus replication
Virology Journal (2024)
-
RNA m6A methylation regulators in liver cancer
Cancer Cell International (2024)
-
M6A plays a potential role in carotid atherosclerosis by modulating immune cell modification and regulating aging-related genes
Scientific Reports (2024)
-
METTL3 facilitates renal cell carcinoma progression by PLOD2 m6A-methylation under prolonged hypoxia
Cell Death & Disease (2024)