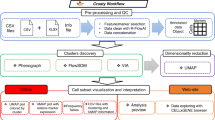
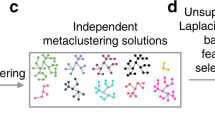
Key Points
-
Recent advances in flow and mass cytometry have spurred the development of novel computational tools to assist in data analysis and visualization. These techniques should be adopted, evaluated and improved upon by the broad immunological community.
-
Standardization is key to making computational flow cytometry work, and researchers should use standard procedures for data generation, analysis, interpretation and deposition. Standardized marker panels should be used as much as possible.
-
Computational flow cytometry allows the automation of population identification, biomarker discovery and predictive modelling to highlight potentially new and interesting cell types that correlate with clinical outcomes.
-
New algorithms allow the modelling of gradual changes that can shed new light on cell developmental processes.
-
Computational flow cytometry offers an additional toolbox, and young immunologists should be trained in basic programming and modelling skills to be able to adequately use these tools and interpret their outcome.
Abstract
Recent advances in flow cytometry allow scientists to measure an increasing number of parameters per cell, generating huge and high-dimensional datasets. To analyse, visualize and interpret these data, newly available computational techniques should be adopted, evaluated and improved upon by the immunological community. Computational flow cytometry is emerging as an important new field at the intersection of immunology and computational biology; it allows new biological knowledge to be extracted from high-throughput single-cell data. This Review provides non-experts with a broad and practical overview of the many recent developments in computational flow cytometry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fulwyler, M. J. Electronic separation of biological cells by volume. Science 150, 910–911 (1965).
Gray, J. W. et al. Chromosome measurement and sorting by flow systems. Proc. Natl Acad. Sci. USA 72, 1231–1234 (1975).
Robinson, J. P. & Roederer, M. Flow cytometry strikes gold. Science 350, 739–740 (2015).
Perfetto, S. P., Chattopadhyay, P. K. & Roederer, M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 4, 648–655 (2004).
Chattopadhyay, P. et al. Toward 40+ parameter flow cytometry. Proc. Congress Int. Soc. Advancement Cytom. Abstr. 2014, 215 (2014).
Bandura, D. R. et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813–6822 (2009).
Nolan, J. P. & Condello, D. Spectral flow cytometry. Curr. Protoc. Cytom. http://www.dx.doi.org/10.1002/0471142956.cy0127s63 (2013).
McGrath, K. E., Bushnell, T. P. & Palis, J. Multispectral imaging of hematopoietic cells: where flow meets morphology. J. Immunol. Methods 336, 91–97 (2008).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Nomura, L., Maino, V. C. & Maecker, H. T. Standardization and optimization of multiparameter intracellular cytokine staining. Cytometry A 73, 984–991 (2008).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 12, 191–200 (2012). This work describes a large-scale effort to standardize flow cytometry data generation and analysis within the Human Immunology Project.
Pachon, G., Caragol, I. & Petriz, J. Subjectivity and flow cytometric variability. Nat. Rev. Immunol. 12, 396–396 (2012).
Gouttefangeas, C. et al. Data analysis as a source of variability of the HLA-peptide multimer assay: from manual gating to automated recognition of cell clusters. Cancer Immunol. Immunother. 64, 585–598 (2015).
Irish, J. Beyond the age of cellular discovery. Nat. Immunol. 15, 1095–1097 (2014).
Spidlen, J. et al. Data file standard for flow cytometry version FCS 3.1. Cytometry A 77, 97–100 (2010).
Spidlen, J., Shooshtari, P. T., Kollmann, R. & Brinkman, R. R. Flow cytometry data standards. BMC Res. Notes 4, 50 (2011).
Spidlen, J., Moore, W. & Brinkman, R. R. ISAC's Gating-ML 2.0 data exchange standard for gating description. Cytometry A 87, 683–687 (2015).
Spidlen, J., Bray, C. & Brinkman, R. R. ISAC's classification results file format. Cytometry A 87, 86–88 (2014).
Spidlen, J. & Novo, D. ICEFormat — the image cytometry experiment format. Cytometry A 81, 1015–1018 (2012).
Schlickeiser, S., Streitz, M. & Sawitzki, B. Standardized multi-color flow cytometry and computational biomarker discovery. Methods Mol. Biol. 1371, 225–238 (2016).
Roederer, M. A proposal for unified flow cytometer parameter naming. Cytometry A 87, 689–691 (2015).
Tung, J. W., Parks, D. R., Moore, W. A., Herzenberg, L. A. & Herzenberg, L. A. New approaches to fluorescence compensation and visualization of FACS data. Clin. Immunol. 110, 277–283 (2004).
Lee, J. A. et al. MIFlowCyt: the minimum information about a flow cytometry experiment. Cytometry A 73, 926–930 (2008).
van Dongen, J. J. M. et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal reactive and malignant leukocytes. Leukemia 26, 1908–1975 (2012). This work describes a community-wide effort to standardize flow cytometry marker panel design for leukaemic disorders.
Finak, G. et al. Standardizing flow cytometry immunophenotyping analysis from the Human ImmunoPhenotyping Consortium. Sci. Rep. 6, 20686 (2016).
Hasan, M. et al. Semiautomated and standardized cytometric procedures for multi-panel and multi-parametric whole blood immunophenotyping. Clin. Immunol. 157, 261–276 (2015).
Mahnke, Y., Chattopadhyay, P. & Roederer, M. Publication of optimized multicolor immunofluorescence panels. Cytometry A 77, 814–818 (2010).
Aghaeepour, N. et al. Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods 10, 445–445 (2013). This work highlights the results of the FlowCAP I and II challenges, which are benchmarks for comparing automated methods for computational flow cytometry.
Aghaeepour, N. et al. A benchmark for evaluation of algorithms for identification of cellular correlates of clinical outcomes. Cytometry A 89, 16–21 (2015).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Perfetto, S. P., Ambrozak, D., Nguyen, R., Chattopadhyay, P. & Roederer, M. Quality assurance for polychromatic flow cytometry. Nat. Protoc. 1, 1522–1530 (2006).
Hahne, F. et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 10, 106 (2009).
Le Meur, N. et al. Data quality assessment of ungated flow cytometry data in high throughput experiments. Cytometry A 71, 393–403 (2007).
Fletez-Brant, K. et al. flowClean: automated identification and removal of fluorescence anomalies in flow cytometry data. Cytometry A 89, 461–471 (2016).
Hahne, F. et al. Per-channel basis normalization methods for flow cytometry data. Cytometry A 77, 121–131 (2010).
Finak, G. et al. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol. 10, e1003806 (2014).
Malek, M. et al. flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics 31, 606–607 (2015).
Kvistborg, P. et al. Thinking outside the gate: single-cell assessments in multiple dimensions. Immunity 42, 591–592 (2015).
Chester, C. & Maecker, H. T. Algorithmic tools for mining high-dimensional cytometry data. J. Immunol. 195, 773–779 (2015).
Mair, F. et al. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur. J. Immunol. 46, 34–43 (2016).
Diggins, K. E., Ferrell, P. B., Irish, J. M. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 82, 55–63 (2015). This work describes an in-depth overview of visualization methods for high-dimensional cytometry data analysis.
Krishnaswamy, S. et al. Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014).
Bendall, S. C. et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014). This work introduces the concept of cell development modelling, showcasing the Wanderlust algorithm applied to B cell development.
Lugli, E. et al. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytometry A 71, 334–344 (2007).
Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Machine Learn. Res. 9, 2579–2605 (2008).
Amir, E. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Cheng, Y., Wong, M. T., van der Maaten, L. & Newell, E. W. Categorical analysis of human T cell heterogeneity with one-dimensional soli-expression by nonlinear stochastic embedding. J. Immunol. 196, 924–932 (2016).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Zunder, E. R., Lujan, E., Goltsev, Y., Wernig, M. & Nolan, G. P. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell 16, 323–337 (2015).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015).
Spitzer, M. H. et al. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425 (2015).
Pyne, S. et al. Automated high-dimensional flow cytometric data analysis. Proc. Natl Acad. Sci. USA 106, 8519–8524 (2009).
Lo, K., Brinkman, R. R. & Gottardo, R. Automated gating of flow cytometry data via robust model-based clustering. Cytometry A 73, 321–332 (2008).
Finak, G., Bashashati, A., Brinkman, R. & Gottardo, R. Merging mixture components for cell population identification in flow cytometry. Adv. Bioinformatics 2009, 247646 (2009).
Chen, X. et al. Automated flow cytometric analysis across large numbers of samples and cell types. Clin. Immunol. 157, 249–260 (2015).
Sorensen, T., Baumgart, S., Durek, P., Grutzkau, A. & Haupl, T. immunoClust — an automated analysis pipeline for the identification of immunophenotypic signatures in high-dimensional cytometric datasets. Cytometry A 87, 603–615 (2015).
Naim, I. et al. SWIFT-scalable clustering for automated identification of rare cell populations in large high-dimensional flow cytometry datasets, part 1: algorithm design. Cytometry A 85, 408–421 (2014).
Aghaeepour, N., Nikolic, R., Hoos, H. H. & Brinkman, R. R. Rapid cell population identification in flow cytometry data. Cytometry A 79, 6–13 (2010).
Zare, H., Shooshtari, P., Gupta, A. & Brinkman, R. R. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics 11, 403 (2010).
Qian, Y. et al. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B. Clin. Cytom 78, S69–S82 (2010).
Ge, Y. & Sealfon, S. C. flowPeaks: a fast unsupervised clustering for flow cytometry data via K-means and density peak finding. Bioinformatics. 28, 2052–2058 (2012).
Johnsson, K., Wallin, J. & Fontes, M. BayesFlow: latent modeling of flow cytometry cell populations. BMC Bioinformatics 17, 25 (2016).
Shekhar, K., Brodin, P., Davis, M. M. & Chakraborty, A. K. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE). Proc. Natl Acad. Sci. USA 111, 202–207 (2013).
Becher, B. et al. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol. 15, 1181–1189 (2014).
Cron, A. et al. Hierarchical modeling for rare event detection and cell subset alignment across flow cytometry samples. PLoS Comput. Biol. 9, e1003130 (2013).
Dundar, M., Akova, F., Yerebakan, H. Z. & Rajwa, B. A non-parametric Bayesian model for joint cell clustering and cluster matching: identification of anomalous sample phenotypes with random effects. BMC Bioinformatics. 15, 314 (2014).
Hsiao, C. et al. Mapping cell populations in flow cytometry data for cross-sample comparison using the Friedman-Rafsky test statistic as a distance measure. Cytometry A 89, 71–88 (2016).
Feher, K., Kirsch, J., Radbruch, A., Chang, H. D. & Kaiser, T. Cell population identification using fluorescence-minus-one controls with a one-class classifying algorithm. Bioinformatics. 30, 3372–3378 (2014).
Zare, H. et al. Automated analysis of multidimensional flow cytometry data improves diagnostic accuracy between mantle cell lymphoma and small lymphocytic lymphoma. Am. J. Clin. Pathol. 137, 75–85 (2012).
Bashashati, A. et al. B cells with high side scatter parameter by flow cytometry correlate with inferior survival in diffuse large B-cell lymphoma. Am. J. Clin. Pathol. 137, 805–814 (2012).
O'Neill, K., Jalali, A., Aghaeepour, N., Hoos, H. & Brinkman, R. R. Enhanced flowType/RchyOptimyx: a Bioconductor pipeline for discovery in high-dimensional cytometry data. Bioinformatics 30, 1329–1330 (2014).
Bruggner, R. V., Bodenmiller, B., Dill, D. L., Tibshirani, R. J. & Nolan, G. P. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl Acad. Sci. USA 111, E2770–E2777 (2014).
Lin, L. et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat. Biotechnol. 33, 610–616 (2015).
Rebhahn, J. A. et al. Competitive SWIFT cluster templates enhance detection of aging changes. Cytometry A 89, 59–70 (2016).
Van Gassen, S., Vens, C., Dhaene, T., Lambrecht, B. N. & Saeys, Y. FloReMi: flow density survival regression using minimal feature redundancy. Cytometry A 89, 22–29 (2016).
Bagwell, C. B. et al. Probability state modeling theory. Cytometry A 87, 646–660 (2015).
Inokuma, M. S., Maino, V. C. & Bagwell, C. B. Probability state modeling of memory CD8+ T-cell differentiation. J. Immunol. Methods 397, 8–17 (2013).
Bagwell, C. B. et al. Human B-cell and progenitor stages as determined by probability state modeling of multidimensional cytometry data. Cytometry B Clin. Cytom. 88, 214–226 (2015).
Marco, E. et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc. Natl Acad. Sci. 111, E5643–E5650 (2014).
Setty, M. et al. Wishbone identifies bifurcating developmental trajectories from single cell data. Nat. Biotechnol. http://dx.doi.org/10.1038/nbt.3569 (2016).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Shin, J. et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015).
Macaulay, I. C. et al. Single-cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep. 14, 966–977 (2016).
Acknowledgements
S.V.G. is funded by the Flanders Agency for Innovation by Science and Technology (IWT). Y.S. is an ISAC Marylou Ingram Scholar. B.N.L. is funded by a European Research Council (ERC) Consolidator grant and several FWO (Research Foundation Flanders) grants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Saeys, Y., Van Gassen, S. & Lambrecht, B. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol 16, 449–462 (2016). https://doi.org/10.1038/nri.2016.56
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri.2016.56
This article is cited by
-
CytoPipeline and CytoPipelineGUI: a Bioconductor R package suite for building and visualizing automated pre-processing pipelines for flow cytometry data
BMC Bioinformatics (2024)
-
Impact of hemodilution on flow cytometry based measurable residual disease assessment in acute myeloid leukemia
Leukemia (2024)
-
Advancing CAR T cell therapy through the use of multidimensional omics data
Nature Reviews Clinical Oncology (2023)
-
BioNetGMMFit: estimating parameters of a BioNetGen model from time-stamped snapshots of single cells
npj Systems Biology and Applications (2023)
-
IMC-Denoise: a content aware denoising pipeline to enhance Imaging Mass Cytometry
Nature Communications (2023)