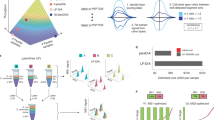
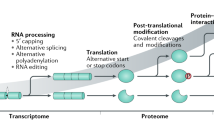
Abstract
Over the past 5 years, protein-chip technology has emerged as a useful tool for the study of many kinds of protein interactions and biochemical activities. The construction of Saccharomyces cerevisiae whole-proteome arrays has enabled further studies of such interactions in a proteome-wide context. Here, we explore some of the recent advances that have been made at the '-omic' level using protein microarrays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ekins, R. P. Multi-analyte immunoassay. J. Pharm. Biomed. Anal. 7, 155–168 (1989).
Ekins, R., Chu, F. & Biggart, E. Multispot, multianalyte, immunoassay. Ann. Biol. Clin. (Paris) 48, 655–666 (1990).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Churchill, G. A. Fundamentals of experimental design for cDNA microarrays. Nature Genet. 32 (Suppl.), 490–495 (2002).
Holloway, A. J., van Laar, R. K., Tothill, R. W. & Bowtell, D. D. Options available — from start to finish — for obtaining data from DNA microarrays II. Nature Genet. 32 (Suppl.), 481–489 (2002).
Quackenbush, J. Microarray data normalization and transformation. Nature Genet. 32 (Suppl.), 496–501 (2002).
Slonim, D. K. From patterns to pathways: gene expression data analysis comes of age. Nature Genet. 32 (Suppl.), 502–508 (2002).
Chuaqui, R. F. et al. Post-analysis follow-up and validation of microarray experiments. Nature Genet. 32 (Suppl.), 509–514 (2002).
Duyk, G. M. Sharper tools and simpler methods. Nature Genet. 32 (Suppl.), 465–468 (2002).
Stoeckert, C. J. Jr, Causton, H. C. & Ball, C. A. Microarray databases: standards and ontologies. Nature Genet. 32 (Suppl.), 469–473 (2002).
MacBeath, G. Protein microarrays and proteomics. Nature Genet. 32 (Suppl.), 526–532 (2002).
Zhu, H. & Snyder, M. Protein chip technology. Curr. Opin. Chem. Biol. 7, 55–63 (2003).
Stoll, D., Templin, M. F., Bachmann, J. & Joos, T. O. Protein microarrays: applications and future challenges. Curr. Opin. Drug Discov. Devel. 8, 239–252 (2005).
Predki, P. F. Functional protein microarrays: ripe for discovery. Curr. Opin. Chem. Biol. 8, 8–13 (2004).
LaBaer, J. & Ramachandran, N. Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 9, 14–19 (2005).
Reboul, J. et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nature Genet. 34, 35–41 (2003).
Rual, J. F. et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004).
Scheich, C., Sievert, V. & Bussow, K. An automated method for high-throughput protein purification applied to a comparison of His-tag and GST-tag affinity chromatography. BMC Biotechnol. 3, 12 (2003).
Ramachandran, N. et al. Self-assembling protein microarrays. Science 305, 86–90 (2004).
Albala, J. S. et al. From genes to proteins: high-throughput expression and purification of the human proteome. J. Cell. Biochem. 80, 187–191 (2000).
Zhu, H. et al. Global analysis of protein activities using proteome chips. Science 293, 2101–2105 (2001).
Goffeau, A. et al. Life with 6000 genes. Science 274, 563–547 (1996).
Gelperin, D. M. et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005).
Angenendt, P. Progress in protein and antibody microarray technology. Drug Discov. Today 10, 503–511 (2005).
Delehanty, J. B. & Ligler, F. S. Method for printing functional protein microarrays. BioTechniques 34, 380–385 (2003).
Pellois, J. P. et al. Individually addressable parallel peptide synthesis on microchips. Nature Biotechnol. 20, 922–926 (2002).
MacBeath, G. & Schreiber, S. L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000).
Newman, J. R. & Keating, A. E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300, 2097–2101 (2003).
Espejo, A., Cote, J., Bednarek, A., Richard, S. & Bedford, M. T. A protein–domain microarray identifies novel protein–protein interactions. Biochem. J. 367, 697–702 (2002).
Kersten, B. et al. Generation of Arabidopsis protein chips for antibody and serum screening. Plant Mol. Biol. 52, 999–1010 (2003).
Lueking, A. et al. A nonredundant human protein chip for antibody screening and serum profiling. Mol. Cell. Proteomics 2, 1342–1349 (2003).
Michaud, G. A. et al. Analyzing antibody specificity with whole proteome microarrays. Nature Biotechnol. 21, 1509–1512 (2003).
Satoh, J. I., Nanri, Y. & Yamamura, T. Rapid identification of 14–3–3-binding proteins by protein microarray analysis. J. Neurosci. Methods 152, 278–288 (2005).
Apweiler, R., Hermjakob, H. & Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999).
Csank, C. et al. Three yeast proteome databases: YPD, PombePD, and CalPD (MycoPathPD). Methods Enzymol. 350, 347–373 (2002).
Hall, D. A. et al. Regulation of gene expression by a metabolic enzyme. Science 306, 482–484 (2004).
Ficarro, S. B. et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nature Biotechnol. 20, 301–305 (2002).
Feilner, T. et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4, 1558–1568 (2005).
The Arabidopsis Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Widmann, C., Gibson, S., Jarpe, M. B. & Johnson, G. L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180 (1999).
Ptacek, J. et al. Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 (2005).
Gavin, A. C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA 98, 4569–4574 (2001).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Xenarios, I. et al. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30, 303–305 (2002).
Bader, G. D. & Hogue, C. W. BIND — a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics 16, 465–477 (2000).
Horak, C. E. et al. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16, 3017–3033 (2002).
Lee, T. I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804 (2002).
Mewes, H. W., Albermann, K., Heumann, K., Liebl, S. & Pfeiffer, F. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 25, 28–30 (1997).
Manning, G., Plowman, G. D., Hunter, T. & Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520 (2002).
Jones, R. B., Gordus, A., Krall, J. A. & MacBeath, G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 (2006).
Kuno, A. et al. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nature Methods 2, 851–856 (2005).
Jin, F. et al. A pooling-deconvolution strategy for biological network elucidation. Nature Methods 3, 183–189 (2006).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
Dr. Snyder is a consultant with Invitrogen.
Related links
Rights and permissions
About this article
Cite this article
Kung, L., Snyder, M. Proteome chips for whole-organism assays. Nat Rev Mol Cell Biol 7, 617–622 (2006). https://doi.org/10.1038/nrm1941
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1941
This article is cited by
-
RNF43 ubiquitinates and degrades phosphorylated E-cadherin by c-Src to facilitate epithelial-mesenchymal transition in lung adenocarcinoma
BMC Cancer (2019)
-
CASCADE_SCAN: mining signal transduction network from high-throughput data based on steepest descent method
BMC Bioinformatics (2011)
-
Proteomics: a pragmatic perspective
Nature Biotechnology (2010)
-
Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes
Molecular Systems Biology (2009)
-
Proteomes take the electrophilic bait
Nature Chemical Biology (2008)