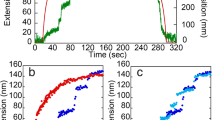
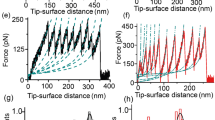
Abstract
Proteins show diverse responses when placed under mechanical stress. The molecular origins of their differing mechanical resistance are still unclear, although the orientation of secondary structural elements relative to the applied force vector is thought to have an important function. Here, by using a method of protein immobilization that allows force to be applied to the same all-β protein, E2lip3, in two different directions, we show that the energy landscape for mechanical unfolding is markedly anisotropic. These results, in combination with molecular dynamics (MD) simulations, reveal that the unfolding pathway depends on the pulling geometry and is associated with unfolding forces that differ by an order of magnitude. Thus, the mechanical resistance of a protein is not dictated solely by amino acid sequence, topology or unfolding rate constant, but depends critically on the direction of the applied extension.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Carrion-Vazquez, M. et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA 96, 3694–3699 (1999).
Oberhauser, A.F., Badilla-Fernandez, C., Carrion-Vazquez, M. & Fernandez, J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319, 433–447 (2002).
Carrion-Vazquez, M. et al. Mechanical design of proteins—studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 74, 63–91 (2000).
Lu, H. & Schulten, K. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins 35, 453–463 (1999).
Klimov, D.K. & Thirumalai, D. Native topology determines force-induced unfolding pathways in globular proteins. Proc. Natl. Acad. Sci. USA 97, 7254–7259 (2000).
Li, H.B., Oberhauser, A.F., Fowler, S.B., Clarke, J. & Fernandez, J.M. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA 97, 6527–6531 (2000).
Brockwell, D.J. et al. The effect of core destabilization on the mechanical resistance of I27. Biophys. J. 83, 458–472 (2002).
Paci, E. & Karplus, M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J. Mol. Biol. 288, 441–459 (1999).
Bryant, Z., Pande, V.S. & Rokhsar, D.S. Mechanical unfolding of a β-hairpin using molecular dynamics. Biophys. J. 78, 584–589 (2000).
Jones, D.D., Stott, K.M., Howard, M.J. & Perham, R.N. Restricted motion of the lipoyl-lysine swinging arm in the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 39, 8448–8459 (2000).
Perham, R.N. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69, 961–1004 (2000).
Zinober, R.C. et al. Mechanically unfolding proteins: the effect of unfolding history and the supramolecular scaffold. Protein Sci. 11, 2759–2765 (2002).
Grandbois, M., Beyer, M., Rief, M., Clausen-Schaumann, H. & Gaub, H.E. How strong is a covalent bond? Science 283, 1727–1730 (1999).
Marko, J.F. & Siggia, E.D. Stretching DNA. Macromolecules 28, 8759–8770 (1995).
Best, R.B., Li, B., Steward, A., Daggett, V. & Clarke, J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 81, 2344–2356 (2001).
Evans, E. & Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 (1997).
Williams, P.M. & Evans, E. In Les Houchesndash;Ecole d'Ete de Physique Theorique Vol. 75, 187–203 (Springer, Heidelberg, Germany, 2002).
Rohs, R., Etchebest, C. & Lavery, R. Unraveling proteins: a molecular mechanics study. Biophys. J. 76, 2760–2768 (1999).
Marszalek, P.E. et al. Atomic levers control pyranose ring conformations. Proc. Natl. Acad. Sci. USA 96, 7894–7898 (1999).
Marszalek, P.E., Li, H.B., Oberhauser, A.F. & Fernandez, J.M. Chair-boat transitions in single polysaccharide molecules observed with force-ramp AFM. Proc. Natl. Acad. Sci. USA 99, 4278–4283 (2002).
Marszalek, P.E. et al. Mechanical unfolding intermediates in titin modules. Nature 402, 100–103 (1999).
Fowler, S.B. et al. Mechanical unfolding of a titin Ig domain: structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J. Mol. Biol. 322, 841–849 (2002).
Lu, H., Isralewitz, B., Krammer, A., Vogel, V. & Schulten, K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75, 662–671 (1998).
Paci, E. & Karplus, M. Unfolding proteins by external forces and temperature: the importance of topology and energetics. Proc. Natl. Acad. Sci. USA 97, 6521–6526 (2000).
Yang, G.L. et al. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. USA 97, 139–144 (2000).
Carrion-Vazquez, M., Li, H., Marszalek, P.E., Oberhauser, A.F. & Fernandez, J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10, 738–743 (2003).
Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J.M. & Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 (1997).
Carl, P., Kwok, C.H., Manderson, G., Speicher, D.W. & Discher, D.E. Forced unfolding modulated by disulfide bonds in the Ig domains of a cell adhesion molecule. Proc. Natl. Acad. Sci. USA 98, 1565–1570 (2001).
Altmann, S.M. et al. Pathways and intermediates in forced unfolding of spectrin repeats. Structure 10, 1085–1096 (2002).
Oberhauser, A.F., Marszalek, P.E., Erickson, H.P. & Fernandez, J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature 393, 181–185 (1998).
Li, F.Y., Yuan, J.M. & Mou, C.Y. Mechanical unfolding and refolding of proteins: an off–lattice model study. Phys. Rev. E 6302, art. no.-021905 (2001).
Rief, M., Pascual, J., Saraste, M. & Gaub, H.E. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286, 553–561 (1999).
Neupert, W. & Brunner, M. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3, 555–565 (2002).
Lee, C., Schwartz, M.P., Prakash, S., Iwakura, M. & Matouschek, A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7, 627–637 (2001).
Lim, J.H., Martin, F., Guiard, B., Pfanner, N. & Voos, W. The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J. 20, 941–950 (2001).
Huang, S.H., Ratliff, K.S., Schwartz, M.P., Spenner, J.M. & Matouschek, A. Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nat. Struct. Biol. 6, 1132–1138 (1999).
Okamoto, K. et al. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 21, 3659–3671 (2002).
Jones, D.D., Stott, K.M., Reche, P.A. & Perham, R.N. Recognition of the lipoyl domain is the ultimate determinant of substrate channelling in the pyruvate dehydrogenase multienzyme complex. J. Mol. Biol. 305, 49–60 (2001).
Brooks, B.R. et al. Charmm—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Lazaridis, T. & Karplus, M. Effective energy function for proteins in solution. Proteins 35, 133–152 (1999).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Murphy, M.E.P. Raster3D Version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure—pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Acknowledgements
We thank A. Berry for help with producing Figure 1 and K. Ainley for the large-scale microbial cultures. We thank A. Blake and the other members of the Radford laboratory for fruitful discussions. We acknowledge the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, University of Leeds, the Wellcome Trust and Forschungskredit der Universität Zürich for financial support. S.E.R. is a BBSRC Professorial Fellow. The manuscript is a contribution from the Astbury Centre for Structural Molecular Biology, which is part of the North of England Structural Biology Centre (NESBIC) and is funded by the BBSRC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Brockwell, D., Paci, E., Zinober, R. et al. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat Struct Mol Biol 10, 731–737 (2003). https://doi.org/10.1038/nsb968
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb968
This article is cited by
-
The role of single-protein elasticity in mechanobiology
Nature Reviews Materials (2022)
-
Detection of weak non-covalent cation-π interactions in NGAL by single-molecule force spectroscopy
Nano Research (2022)
-
Distal conformational locks on ferrocene mechanophores guide reaction pathways for increased mechanochemical reactivity
Nature Chemistry (2021)
-
Anisotropy in mechanical unfolding of protein upon partner-assisted pulling and handle-assisted pulling
Communications Biology (2021)
-
Protein folding modulates the chemical reactivity of a Gram-positive adhesin
Nature Chemistry (2021)