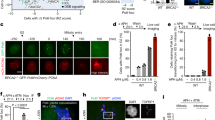
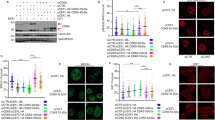
Abstract
Homologous recombination (HR) and nonhomologous end joining (NHEJ) are two distinct DNA double-stranded break (DSB) repair pathways. Here, we report that DNA-dependent protein kinase (DNA-PK), the core component of NHEJ, partnering with DNA-damage checkpoint kinases ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR), regulates HR repair of DSBs. The regulation was accomplished through modulation of the p53 and replication protein A (RPA) interaction. We show that upon DNA damage, p53 and RPA were freed from a p53–RPA complex by simultaneous phosphorylations of RPA at the N-terminus of RPA32 subunit by DNA-PK and of p53 at Ser37 and Ser46 in a Chk1/Chk2-independent manner by ATR and ATM, respectively. Neither the phosphorylation of RPA nor of p53 alone could dissociate p53 and RPA. Furthermore, disruption of the release significantly compromised HR repair of DSBs. Our results reveal a mechanism for the crosstalk between HR repair and NHEJ through the co-regulation of p53–RPA interaction by DNA-PK, ATM and ATR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S . Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73: 39–85.
Huen MS, Chen J . Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 2009; 35: 101–108.
Harper JW, Elledge SJ . The DNA damage response: ten years after. Mol Cell 2007; 28: 739–745.
Wood JL, Chen J . DNA-damage checkpoints: location, location, location. Trends Cell Biol 2008; 18: 451–455.
Zou Y, Liu Y, Wu X, Shell SM . Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol 2006; 208: 267–273.
Shell SM, Zou Y . Protein-protein interactions in Ataxia telangiectesia. In: Ahmad S, Hanaoka F (eds). Molecular Mechanisms of Ataxia Telangiectasia. Landes Bioscience, Austin, TX, USA, 42–51 2009.
Arnaudeau C, Lundin C, Helleday T . DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol 2001; 307: 1235–1245.
Pommier Y . Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 2006; 6: 789–802.
Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev 2004; 18: 1293–1304.
Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW . Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev 2004; 18: 1283–1292.
Hill R, Lee PW . The DNA-dependent protein kinase (DNA-PK): more than just a case of making ends meet? Cell Cycle 2010; 9: 3460–3469.
Meek K, Dang V, Lees-Miller SP . DNA-PK: the means to justify the ends? Adv Immunol 2008; 99: 33–58.
Wu X, Yang Z, Liu Y, Zou Y . Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem J. 2005; 391: 473–480.
Wu X, Shell SM, Zou Y . Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 2005; 24: 4728–4735.
Zou L, Liu D, Elledge SJ . Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 2003; 100: 13827–13832.
Zou L, Elledge SJ . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300: 1542–1548.
Yang ZG, Liu Y, Mao LY, Zhang JT, Zou Y . Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry 2002; 41: 13012–13020.
Robison JG, Bissler JJ, Dixon K . Replication protein A is required for etoposide-induced assembly of MRE11/RAD50/NBS1 complex repair foci. Cell Cycle 2007; 6: 2408–2416.
Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ et al. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 2000; 103: 449–456.
Sugiyama T, Kantake N . Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. J Mol Biol 2009; 390: 45–55.
Stauffer ME, Chazin WJ . Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J Biol Chem 2004; 279: 25638–25645.
Jackson D, Dhar K, Wahl JK, Wold MS, Borgstahl GE . Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J Mol Biol 2002; 321: 133–148.
Sommers JA, Sharma S, Doherty KM, Karmakar P, Yang Q, Kenny MK et al. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res 2005; 65: 1223–1233.
Romanova LY, Willers H, Blagosklonny MV, Powell SN . The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene 2004; 23: 9025–9033.
Derheimer FA, O’Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, RPA Ljungman M . and ATR link transcriptional stress to p53. Proc Natl Acad Sci USA 2007; 104: 12778–12783.
Hass CS, Gakhar L, Wold MS . Functional characterization of a cancer causing mutation in human replication protein A. Mol Cancer Res 2010; 8: 1017–1026.
Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 2005; 37: 750–755.
Binz SK, Sheehan AM, Wold MS . Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 2004; 3: 1015–1024.
Vassin VM, Wold MS, Borowiec JA . Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol 2004; 24: 1930–1943.
Patrick SM, Oakley GG, Dixon K, Turchi JJ . DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry 2005; 44: 8438–8448.
Guo S, Zhang Y, Yuan F, Gao Y, Gu L, Wong I et al. Regulation of replication protein A functions in DNA mismatch repair by phosphorylation. J Biol Chem 2006; 281: 21607–21616.
Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y . Modulation of replication protein A function by its hyperphosphorylation-induced conformational change involving DNA binding domain B. J Biol Chem 2005; 280: 32775–32783.
Shi W, Feng Z, Zhang J, Gonzalez-Suarez I, Vanderwaal RP, Wu X et al. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis 2010; 31: 994–1002.
Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X . RPA2 Is a Direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem 2006; 281: 39517–39533.
Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K et al. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry 2005; 44: 8428–8437.
Vise PD, Baral B, Latos AJ, Daughdrill GW . NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res 2005; 33: 2061–2077.
Bochkareva E, Kaustov L, Ayed A, Yi G-S, Lu Y, Pineda-Lucena A et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA 2005; 102: 15412–15417.
Miller S, Moses K, Jayaraman L, Prives C . Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single- stranded DNA. Mol Cell Biol 1997; 17: 2194–2201.
Abramova NA, Russell J, Botchan M, Li R . Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc Natl Acad Sci USA 1997; 94: 7186–7191.
Kuhn C, Muller F, Melle C, Nasheuer HP, Janus F, Deppert W et al. Surface plasmon resonance measurements reveal stable complex formation between p53 and DNA polymerase alpha. Oncogene 1999; 18: 769–774.
Dutta A, Ruppert JM, Aster JC, Winchester E . Inhibition of DNA replication factor RPA by p53. Nature 1993; 365: 79–82.
Levine AJ . p53, the cellular gatekeeper for growth and division. Cell 1997; 88: 323–331.
Ko LJ, Prives C . p53: puzzle and paradigm. Genes Dev 1996; 10: 1054–1072.
Efeyan A, Serrano M . p53: guardian of the genome and policeman of the oncogenes. Cell Cycle 2007; 6: 1006–1010.
Kaustov L, Yi GS, Ayed A, Bochkareva E, Bochkarev A, Arrowsmith CH . p53 transcriptional activation domain: a molecular chameleon? Cell Cycle 2006; 5: 489–494.
Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y . Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. Embo J 1999; 18: 1397–1406.
Oakley GG, Patrick SM, Yao J, Carty MP, Turchi JJ, Dixon K . RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 2003; 42: 3255–3264.
Sakasai R, Teraoka H, Tibbetts RS . Proteasome inhibition suppresses DNA-dependent protein kinase activation caused by camptothecin. DNA Repair 2009; 9: 76–82.
Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC . The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J Biol Chem 2003; 278: 23410–23417.
Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC . Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. The EMBO J 2006; 25: 5539–5548.
Plate I, Hallwyl SC, Shi I, Krejci L, Muller C, Albertsen L et al. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J Biol Chem 2008; 283: 29077–29085.
Pierce AJ, Hu P, Han M, Ellis N, Jasin M . Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001; 15: 3237–3242.
Johnson RD, Liu N, Jasin M . Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 1999; 401: 397–399.
Shieh SY, Ikeda M, Taya Y, Prives C . DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91: 325–334.
Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet 1998; 20: 398–400.
Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 1998; 281: 1677–1679.
Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 1999; 13: 152–157.
Chehab NH, Malikzay A, Appel M, Halazonetis TD . Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 2000; 14: 278–288.
Virshup DM, Wold MS, Weinberg DH, Kauffman MG, Kelly TJ . Cellular proteins involved in SV40 DNA Replication In vitro. In: Richardson CC, Lehman IR, Alan RL (eds). Molecular Mechanisms in DNA Replication and Recombination. American Society for Microbiology, New York,, 303–314 1990.
Leiter LM, Chen J, Marathe T, Tanaka M, Dutta A . Loss of transactivation and transrepression function, and not RPA binding, alters growth suppression by p53. Oncogene 1996; 12: 2661–2668.
Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM . The antioxidant function of the p53 tumor suppressor. Nat Med 2005; 11: 1306–1313.
Sun J, Oma Y, Harata M, Kono K, Shima H, Kinomura A et al. ATM modulates the loading of recombination proteins onto a chromosomal translocation breakpoint hotspot. PLoS One 2010; 5: e13554.
Anantha RW, Sokolova E, Borowiec JA . RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc Natl Acad Sci USA. 2008; 105: 12903–12908.
Acknowledgements
We gratefully acknowledge Dr Xiaohua Wu for providing U2OS cells expressing RPA32-WT and PD-RPA proteins. We also gratefully acknowledge Dr Carl W Anderson for providing the p53-expression constructs (pCAG3.1-WT, -S15A, -S20A, -S37A and -S46A) and Dr Karen Vousden for the pCB6 expression vectors p53-WT and p53-S15A. This work is supported by National Institutes of Health grants CA86927 and GM083307 (to YZ) as well as ES017214 (to MAS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Serrano, M., Li, Z., Dangeti, M. et al. DNA-PK, ATM and ATR collaboratively regulate p53–RPA interaction to facilitate homologous recombination DNA repair. Oncogene 32, 2452–2462 (2013). https://doi.org/10.1038/onc.2012.257
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.257
Keywords
This article is cited by
-
p53 isoforms differentially impact on the POLι dependent DNA damage tolerance pathway
Cell Death & Disease (2021)
-
CRL4ADTL degrades DNA-PKcs to modulate NHEJ repair and induce genomic instability and subsequent malignant transformation
Oncogene (2021)
-
In vitro dexamethasone treatment does not induce alternative ATM transcripts in cells from Ataxia–Telangiectasia patients
Scientific Reports (2020)
-
Deciphering p53 dynamics and cell fate in DNA damage response using mathematical modeling
Genome Instability & Disease (2020)
-
Radiotherapy and Glioma Stem Cells: Searching for Chinks in Cellular Armor
Current Stem Cell Reports (2017)