Abstract
Brain injury is one of the most consequential problems facing neonates, with many preterm and term infants at risk for cerebral hypoxia and ischemia. To develop effective neuroprotective strategies, the mechanistic basis for brain injury must be understood. The fragile state of neonates presents unique research challenges; invasive measures of cerebral blood flow and oxygenation assessment exceed tolerable risk profiles. Near-infrared spectroscopy (NIRS) can safely and non-invasively estimate cerebral oxygenation, a correlate of cerebral perfusion, offering insight into brain injury-related mechanisms. Unfortunately, lack of standardization in device application, recording methods, and error/artifact correction have left the field fractured. In this article, we provide a framework for neonatal NIRS research. Our goal is to provide a rational basis for NIRS data capture and processing that may result in better comparability between studies. It is also intended to serve as a primer for new NIRS researchers and assist with investigation initiation.
Similar content being viewed by others
Introduction
Brain injury is an important and consequential problem faced by many infants in the neonatal intensive care unit (NICU). Understanding injury mechanisms and developing tools for early intervention to enhance neuroprotection are high priorities. Near-infrared spectroscopy (NIRS) is a promising technology that allows noninvasive assessment of cerebral oxygenation on a bedside monitor [1, 2]. It has been widely adopted over the past two decades for research and is now increasingly being deployed for clinical use [3,4,5].
However, the use of NIRS to monitor regional tissue oxygenation in neonates remains a widely fractured field. This is likely a consequence of absent standards and coordination of the various stages of NIRS monitoring, including data capture, processing, and methods for assessing oxygenation, oxygen extraction, and autoregulation. For autoregulation specifically, there have been numerous studies with concordant findings, but significant differences in approach make comparing trials a daunting task [6,7,8,9,10].
In this manuscript, we provide an overview of NIRS monitoring in the neonate, approaches for capturing data from commonly used devices, data handling methods, and techniques to assess NIRS-based cerebral autoregulation. These guidelines are intended for use in the neonatal population and cover a broad range of equipment and software. We conclude with a set of recommended practices, with the aim of providing a foundation for future neonatal NIRS practices.
Equipment
NIRS fundamentals
NIRS is a non-invasive technique for measuring the percentage of saturated hemoglobin in a target tissue. It relies on two physical principles: differential absorption of near-infrared light and the modified Beer–Lambert law [11]. NIRS devices utilize light in the near-infrared band (700–900 nm), to which skin, bone, and connective tissue are mostly transparent [12, 13]. As NIR light diffuses into the tissue, it interacts with hemoglobin in four different ways: absorption, reflection, scattering, and transmission [13]. Although transmitted and absorbed light are lost and not returned to the sensor, a portion of the source light is reflected back to the sensor, and a smaller portion is scattered by motion, generally, blood flowing through arteries. Over the time frame of a typical NIRS recording, the underlying structure of the monitored tissue remains constant, therefore reflection, transmission, and scattering are assumed to be constant as well.
Thus, the only variable optical factor is absorption, which changes based on the degree of oxygen saturation. The absorption spectra of oxy- and deoxyhemoglobin in the near-infrared band are different, with greater absorption by deoxyhemoglobin (Fig. 1). Light of at least two different wavelengths (above and below 810 nm) is applied to tissues of interest, and the relative concentration of oxygenated and deoxygenated hemoglobin is estimated using the modified Beer–Lambert law, thus providing an index of tissue oxygenation [14]. Experimental NIRS systems utilize other optical properties and wavelengths of light as detailed below.
Unlike pulse oximetry, NIRS measurements are not pulse-synchronized and thus not limited to arterial hemoglobin sources. Instead, the light interrogates arterial, venous, and capillary beds, providing a regionalized composite measure. Given that only ~30% of blood is intraarterial at any given time in most tissues, NIRS monitoring provides an approximate 30/70 arterial/venous-weighted estimate of oxygen saturation [15, 16] which closely parallels jugular venous oxygen saturation [17].
Commercial NIRS monitors
A range of commercially available NIRS monitors with neonatal indications is listed in Table 1. Although all NIRS monitors operate under the fundamentals described previously, each contains proprietary signal-processing algorithms. While this systematic difference in measured saturation precludes comparison of absolute measurements, head-to-head comparison (NIRO vs. INVOS [18, 19], Nonin vs. ForeSight [20], INVOS vs. ForeSight [21]) of competing devices demonstrates the generally strong correlation between devices, supporting the notion that, despite proprietary differences, they are largely equivalent. Further, recent publications by Kleiser using a blood-lipid phantom have mathematically modeled the differences between devices, allowing for precise correction of measurements between devices [22,23,24].
Research monitors
Although there is some variation in emitter-detector difference and wavelength selection, all commercial NIRS devices use a single probe to detect regional oxygenation. Several different research optical devices are currently in development which greatly extend this technology into new avenues and may provide valuable additional information beyond regional tissue oxygenation.
One such technique is diffuse correlation spectroscopy (DCS). While currently available commercial NIRS devices utilize only differences in light absorption between oxyhemoglobin and deoxyhemoglobin, DCS also detects the scattering of light from moving red blood cells, enabling measurement of cerebral blood flow in addition to saturation [25]. DCS systems have been validated for neonates in a number of studies [26, 27], including infants with congenital heart disease [28]. In a recent publication, the prototype BabyLux commercial DCS system was detailed [29].
Another technology, functional NIRS (fNIRS) or the more advanced derivative diffuse optical tomography (DOT), utilizes 10–20 or more individual optical channels embedded in “caps” with fixed optode positions. By synchronized capture of all channels, the system reconstructs three-dimensional cerebral oxygenation patterns across large regions of the cerebral cortex [30]. In this manner, changes in regional oxygenation in the setting of injury or response to stimuli can reveal metabolically connected networks, similar to functional MRI [31, 32]. In neonates, DOT has been used to classify the hemodynamic response during seizures [33] and for the detection of intraventricular hemorrhage [34].
Finally, broadband NIRS is a relatively new technique that has recently been applied to neonates. Unlike conventional NIRS which utilizes 3–5 fixed wavelengths of light, broadband NIRS utilizes more than 100 frequencies of light to characterize cytochrome-c-oxidase (CCO), the terminal electron acceptor in the electron transport chain [35]. Changes in the oxidation state of CCO can be detected using broadband NIRS and are thought to be more reflective of brain metabolism than oxy-/deoxyhemoglobin concentration [36]. Given the small concentration of CCO in the brain and confounding by competing for absorption by hemoglobin, CCO measurement has only recently become practical. Broadband NIRS has been demonstrated in piglet models of HIE [37] as well as pilot studies of human neonates with HIE [35, 38].
Probes
The majority of commercial NIRS monitors have specific pediatric or neonatal-sized sensors available. Beyond physical size differences from the adult probes, pediatric and neonatal probes also have shorter distances between the emitting light source and the detectors, altering the depth of penetration to provide greater sensitivity to signals transmitted through the thinner skull of an infant [39]. Differences in tissue saturation values between adult, pediatric, and neonatal sensors may range between 10% and 14% [20]. In addition, reference values for cerebral oxygenation measures in preterm infants have been based on studies using small adult sensors [40, 41], with the limited investigation into the optimal adjustment required for neonatal sensors [20].
The selection of a NIRS device is an institutional decision beyond the scope of this report. Key factors to consider include: device and sensor costs, availability of adhesive and non-adhesive probes (particularly important for preterm infants), availability of single-use or reusable probes, ease of cleaning for infection control, regional regulatory approval (allowing research and clinical use on the same devices), and compatibility with existing monitoring hardware. A non-exhaustive search of prospective cerebral oxygenation studies reveals that the bulk of neonatal NIRS research has been conducted with the INVOS (Medtronic, Minneapolis, MN, USA), NIRO (Hamamatsu Photonics, Hamamatsu City, Shizuoka, Japan), and ForeSight (Edwards Lifesciences, Irvine, CA, USA) devices.
Data capture options
Commercially available NIRS devices are primarily intended for clinical use with a focus on trend monitoring and not long-term analysis. As such, recording data is a secondary consideration that requires additional planning, infrastructure, and costs. Key fundamentals of data capture platforms include (a) synchronized capture of comprehensive vital signs, (b) accurate time/date information, and (c) protection against data loss. Potential data capture approaches, including advantages and disadvantages, are presented as follows. Regardless of the method used, data should be captured at the highest possible sampling rate [42].
Local device
The simplest approach to capturing NIRS data is a direct download from NIRS devices. Several manufacturers provide a USB interface for this purpose, including the Hamamatsu, INVOS, and ForeSight devices. While this approach is the most straightforward, it has several drawbacks. First, data can only be downloaded after a monitoring session is complete. Power interruption or accidental deletion from the device can result in data loss, and the limited memory capacity on each device may result in data files being overwritten if not routinely downloaded. Second, this approach only permits the capture of NIRS data; other vital signs are not included in the file. Thus, data captured in this manner needs to be externally harmonized with other vital sign information. Finally, the time stamp for this NIRS data originates from the NIRS device clock, which is frequently not adjusted for daylight savings time or maybe reset in the case of power loss.
Central-hub and central-server data capture
To coordinate the synchronized capture of physiologic signals from multiple sources, a central computer is needed to receive the data and store it in a single, aggregated file. The typical “central-hub” strategy utilizes a laptop computer at the patient’s bedside which is physically connected to the NIRS monitor, the patient monitor, the ventilator, or a host of other devices. This approach is advantageous in that the computer provides the timing data, so only one clock must be checked before starting the recording and the software writes the data directly to disk so that if there is a power interruption, previously recorded data is not lost. Some software packages also include integrated analytic tools, providing a low-resistance path for new investigators. The most significant disadvantage to the “central-hub” model is that generally requires the additional purchase of hardware (laptops, computer cabling) or software and may impose restrictions with limited device compatibility.
A variation on this method for data capture is the “central-server” model, where individual monitoring devices connect over a network to a centrally located server. The approach shares the same advantage of synchronized timing and adds the advantage of mostly automated capture, requiring minimal input from users to start and stop the recording and no need for a laptop at each patient’s bedside. The significant disadvantage is that it is the most complex solution, from an infrastructure standpoint, often requiring significant capital expense.
Software packages from NIRS manufacturers, as well as third-party software and hardware vendors, are listed in Table 2. Examples of INVOS and ForeSight capture configurations are shown in Fig. 2.
Recommendations for data capture
To analyze cerebrovascular hemodynamic data, important steps must be taken in the organization and format of captured data. Standardized data formatting will allow for easy exchange in multi-center collaborative efforts, the development of improved analytic tools, and the elimination of unnecessary conversion work. Three data storage principles are important to consider—open data format, consistent time stamping, and synchronization.
Open format
As previously noted, some software/hardware combinations result in files in a proprietary format. This is disadvantageous for several reasons. First, technological advances will eventually render most formats obsolete, requiring maintaining legacy systems and/or conversion tools to allow continued use of files originating in these systems. Second, proprietary formats may prevent sharing with other investigators lacking the same platform. Third, corruption of data files may result in data lossless manageable than using nonproprietary systems.
In contrast, open file formats, notably comma-separated value (CSV), are not proprietary or platform-specific and can be accessed using a wide range of tools. Processing tools to summarize data and validate data integrity can be more widely applied with open formats. In addition, CSV files are also tolerant of corruption; although there will be data loss in the corrupted region of the file, the remainder of the file is usually accessible. However, CSV is not a particularly efficient format, resulting in significantly larger files than other formats. However, given the low relative costs of file storage, this has become less of a concern.
Timestamping
Accurate time stamping is essential for quantitative analysis of NIRS signals. There are several approaches to recording time data that may apply in different scenarios. The most ideal approach records to date and time information in an unambiguous fashion. One approach is to utilize the format published by the International Organization for Standards like ISO 8601 [43] which represents dates as YYYY-MM-DD where yyyy is a four-digit year, mm is the two-digit month, and dd is the two-digit date. ISO 8601 also includes a standard format for time representation, hh:mm:ss, where hh represents hours on a 24-h clock, mm is minutes, and ss is seconds.
An alternative approach is to use Unix timestamps, which is a system for describing time as the number of seconds which have elapsed since the “Unix epoch,” 00:00:00 Jan 1, 1970, UTC, accounting for leap seconds [44]. As a decimal value, Unix time-stamp data is easy to utilize for date/time manipulations as it is an integer value such as 1584316920. A distinct disadvantage of Unix time is the loss of daylight saving or time zone information, as all time is referenced to UTC, the successor to Greenwich Mean Time, and is not adjusted for daylight saving. Careful attention must be paid when comparing local time events (such as time of birth, medication administration, etc.) to data recorded in Unix time to ensure the correct offset is applied for locale and DST.
A consistent approach to time stamps should be an achievable priority for the NIRS research community.
Safety and privacy
Digital recordings offer significant convenience and are easily transferred between investigators facilitating collaborative research. This ease of use is also the source of potential safety and privacy concerns. Physiologic recordings are patient medical records and are subject to privacy laws such as HIPAA, PIPEDA, and GDPR. To maintain compliance with these regulations, all recordings should be de-identified, encrypted, and stored in locations with access controls (requiring a login that can be audited) and not on USB flash drives or external hard drives [45].
For files that are shared with other investigators, researchers may wish to remove identifiable dates and times. One approach is to shift the date or time by a fixed amount of time. Alternatively, the time column could be recalculated, for example as the number of seconds since the start of the recording or the number of seconds since birth [46]. However, once recalculated, it is no longer possible to cross-reference time information with external events (such as the initiation of a medication or a known clinical event). In addition, serial time numbers should be recorded in actual units of time (i.e., seconds) so that the sampling rate can be ascertained by others.
Synchronization
Cerebral oximetry measures alone are not sufficient to calculate cerebrovascular autoregulatory function. At least one other source of information (e.g., arterial blood pressure, heart rate, or pulse oximetry) must be used. These other data streams can be used for autoregulatory assessment in addition to error detection and/or correction. However, synchronized recording of these multiple signals is essential for this process to work correctly.
As noted earlier, there are two ways to address this challenge. One approach is to synchronize all monitoring signals into a central monitoring device and sample data directly from the central device. The use of IntelliBridge modules to connect peripheral monitors to a Philips patient monitor is an example of this approach. Alternatively, a bedside computer (or networked server) could serve as the hub for multiple devices. Each device will need to have a physical connection (via serial or network cable) to the hub computer. The Medtronic VitalSync and CAS/Edwards FS DAQ software packages are examples of this approach. A third, significantly less ideal option can be used when separate recording systems are used for each device. In this scenario, a “time mark” can be entered as an annotation in the recording stream. After the recording is complete, each individual file can be combined into a composite file, aligned by the “time mark” point. For this approach to work, it requires the user to press a button on two or more devices at exactly the same time and also requires each device clock to be precisely synchronized.
Approaches to error correction
Physiologic data collected in the NICU is contaminated with signal noise and artifacts. The challenge for researchers is to deploy artifact or error correction tools that reduce the impact of these outliers without accidentally removing valid data or creating significant gaps. Although some methods for cerebrovascular modeling tolerate some degree of missing data, large gaps remain problematic. Artifacts can be divided into three distinct categories: missing values, out of range values, and motion artifact.
Missing values
Missing data is most often caused when a sensor has been disconnected from a patient or the device has been placed in standby mode. For analytic purposes, missing values should not be replaced with zeros, as this numerical value will alter statistical calculations (such as mean or standard deviation) or frequency-domain analysis. Missing values should instead be replaced with a “not a number” (NaN) representation. This approach prevents inadvertent mathematic skewing of results.
Out of range values
Although most values captured during recording will fall within expected normal ranges, some measurements will be outliers. Outlier values can be defined as point outliers, values that are so far outside the normal range that it is clear they are erroneous (e.g., heart rate of 475 bpm), and contextual outliers, which may be of nominally acceptable value but out of place when considering measurements obtained before and after that point [47]. The first type of outlier is considered an “out of range” value which should be removed from the recording. The second type of outlier is the result of motion artifact and will be discussed separately.
When removing out of range values, determination of upper and lower limits should be made a priori utilizing gestational age-appropriate values. While truly normative values are not well established in preterm neonates, existing retrospective data should be sufficient for blood pressure [48,49,50] and heart rate [51]. Point outliers represent error conditions and depart significantly from normal values and a simple screening for values two or three standard deviations from the mean is likely sufficient to exclude them without removing true patient data.
One commonly encountered scenario is exceedingly high or low values for arterial blood pressure, which occur when the line is being accessed for sampling or during infusions. Another common scenario is the generation of error codes, typically significantly out of range values reported by a monitor when a sensor has encountered an error (usually when the sensor is off the patient). Finally, some NIRS monitors have fixed ranges. For example, the INVOS 5100c does not report oxygen saturations below 15%. It is important that clinicians and researchers be aware of specific patient- and monitor-specific circumstances to determine whether out of range data represent a physiologic disturbance vs. a monitoring error. In the latter case, these values can be readily identified as outliers in the recording and can be programmatically removed by replacement of all non-physiologic values with NaN.
Motion artifact
NIRS signals are prone to changes from baseline when patients move, an unavoidable reality in the neonatal population. Sudden signal changes can be recognized as contextual outliers but are more difficult to remove programmatically as the values may still remain within normative ranges. Analysis, therefore, must include methods for detecting and removing motion artifacts. NIRS probes with accelerometer-based movement detection sensors have been described in previous publications [52, 53], but this technology is not yet commercially available.
Fortunately, movement artifacts can also be determined using characteristic changes in the signal. A number of different approaches have been used in prior research including principal component analysis [54, 55], wavelet analysis [56], Kalman filtering [57], sliding window averaging [58], statistical inference [59], non-parametric threshold detection [60], or a combination of multiple approaches [61]. The systematic comparison of the different techniques suggests that the wavelet approach results in the greatest reduction in error, although these comparisons did not include all techniques nor are they neonate-specific [62,63,64]. Regardless of the method, all reports indicate that motion artifact correction is an essential component of data processing.
Interpolation
Interpolation methods refer to “best guesses” about missing values using nearby valid data. Common approaches to vital sign interpolation include nearest-neighbor [65], linear interpolation [66], and spline interpolation [67] (Table 3). The accuracy of these methods to estimate missing data is directly related to the length of missing data and the complexity of the approach used. The systematic comparison of interpolation methods suggests that spline interpolation provides the largest reduction in mean-square error [63, 68]. An example of the output of several approaches to interpolation is shown in Fig. 3.
There is considerable debate about the validity of using interpolation or imputation to fill in gaps caused by missing data [69, 70]. Although these techniques may permit the analysis of otherwise unusable data, this approach results in the mixing of synthetically generated data with real measurements and care must be taken to avoid the introduction of error. Missing data are common in longitudinal analysis and interpolation or imputation is an accepted approach [71]. The success of interpolation techniques is dependent on many factors including the predictability of the signal, the sampling rate, and the length of the missing segment [72]. Large segments of missing data present a difficult challenge that, even with sophisticated techniques, affects many domains of quantitative analysis [73]. Importantly, an empiric study of tolerance to missingness has not been conducted and remains an important unanswered question.
Some researchers have chosen to take a strict approach to missing data, excluding all segments with missing values. This approach does avoid the need for interpolation, but significant exclusion of underlying data may also introduce bias. The percent of missing data during the monitoring period should be reported in these cases. When interpolation is used, it is prudent to additionally report on the same data set without interpolative methods as a form of quality assurance.
Approaches to cerebral autoregulation
Cerebral autoregulation is the mechanism that modulates blood flow to the brain. This complex mechanism is responsible for fine adjustment of blood flow to match delivered oxygen to metabolic demand in the setting of fluctuations induced by changes in cardiac output, carbon dioxide, and physical positioning (e.g., supine vs. upright). Significant evidence exists that autoregulatory function is altered by the degree of prematurity [7] and severity of illness for preterm infants [9] and the degree of brain injury for term infants with HIE [74, 75]. The primary challenge of autoregulation research is the difficulty in direct measurement. Though methods do exist to assess autoregulatory capacity, they are either impractical or unsafe for use in neonates owing to the need for intracranial pressure monitoring, ionizing radiation, and/or radioisotopes.
Far more feasible is the mathematical modeling of the autoregulatory function using known inputs (heart rate, arterial blood pressure) and known outputs (cerebral blood flow—either directly using DCS or indirectly by conventional NIRS oximetry, previously validated as a reliable proxy measurement) [2, 76]. Success in this line of investigation arises from careful planning; the analytic approach informs the data capture strategy as much as the capture approach drives analytics. There is no standard methodology for the characterization of cerebral autoregulation in neonates and several different approaches have been developed.
Time correlation
The simplest and most straightforward approach is to correlate mean arterial blood pressure (MABP) and NIRS measurements. When cerebral autoregulation is intact, there should be a minimal or negative correlation, indicating active autoregulation. To reduce the impact of the non-linear relationship between cerebral blood flow and autoregulation [77], correlations should be calculated over short time windows, most commonly in 20-min segments. Inherent in this approach is the need for a pre-defined threshold beyond which autoregulation is considered impaired. The number of segments with a correlation greater than 0.5 can be summed, and the cumulative number of these “pressure passive” time periods has been linked with adverse outcomes [6].
Time correlation is relatively robust to small segments of missing data. As the typical 20-min window contains 300–600 measurements (depending on sampling rate), the degree of correlation can still be ascertained if some data are missing. However, as the number of missing data increases, per-window estimates become less accurate. An example of this approach is shown in Fig. 4A.
Cerebrovascular reactivity
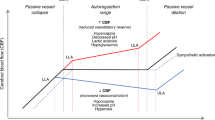
A variation of the standard time-correlation approach, a method measuring cerebrovascular reactivity can be used to identify the “optimal blood pressure.” This approach was initially developed using transcranial Doppler ultrasound [78, 79] but has been more recently adapted to the neonatal population using NIRS [8, 80,81,82,83,84,85]. In this approach, cerebral NIRS and MABP values are captured simultaneously and the correlation between the two is calculated over 5-min overlapping windows. The value of the correlation (termed COx) and the average MABP during the 5-min window are recorded and organized into “bins” of 1–2 mmHg. In this way, the average COx or correlation at each blood pressure can be assessed. Using this approach, one would expect a plot of mean COx by MABP to resemble a parabola, with increasingly positive values at the two ends representing failing autoregulation at BP extremes. In this approach, the “optimal” MABP is defined as the MABP with the lowest mean COx [84].
Cerebrovascular reactivity is quite robust in the presence of missing data owing to shorter time windows (5 min) which can accommodate gaps in the data. Furthermore, in the final stage of calculations, the correlation values in each blood pressure “bin” are averaged across the entire recording. Sufficiently long recordings will have adequate data for a representative sample. An example of this approach is shown in Fig. 4B.
Frequency correlation
A related technique examines the correlation between oscillations of different frequencies in MABP and NIRS. Complex physiologic signals can be thought of as numerous interposed sine waves. The power spectrum, or the strength of oscillations in discrete frequency bands, can be mathematically determined using a technique called the Fourier transform and is performed rapidly using modern computers [86]. As in the time-domain analysis, these oscillations should be dampened by cerebrovascular autoregulation. Strong oscillations in cerebral blood flow (using NIRS as a proxy estimate), at the same frequencies as systemic blood flow, indicate failed autoregulation.
As with the time-domain approach, the recording can be partitioned into blocks of 20 min in which the coherence can be measured. Coherence is a measure of correlation but at a specific frequency or band of frequencies. Unlike a correlation coefficient, values range between 0 and 1, with 1 representing perfect coherence. As with the time-domain technique, coherence requires an agreed-upon threshold, beyond which autoregulation is judged to be impaired. Thresholds for significance have ranged from 0.384 [75] to 0.77 [9] in studies of preterm infants [9, 10, 87, 88] and term infants with hypoxic-ischemic encephalopathy [74, 75].
Calculation of the power spectrum is conventionally performed with Welch’s method and requires continuous, uninterrupted data. Even a single missing point results in an error. The use of data windows allows for the calculation of coherence around missing time points by dropping those windows with missing data. Alternative methods have been developed to determine the power spectrum in the setting of missing data, but require special implementation [89, 90]. An example of coherence calculated over the length of a recording is shown in Fig. 4C.
Transfer function
An alternative frequency-domain approach models autoregulation as a “black box” that filters out blood pressure oscillations at specific frequencies. Autoregulation should serve as a high-pass filter [91] which filters out low-frequency (<0.20 Hz) oscillations in blood pressure, but not fast (high-frequency) changes which are likely needed to respond to rapid changes in cerebral metabolism.
In this method, the simultaneously collected MABP and NIRS values are partitioned into 20-min windows. A well-functioning autoregulatory system, operating as a high-pass filter, should provide significantly stronger dampening at lower frequencies (Fig. 4D). As with cerebrovascular reactivity, this approach was initially developed [92] using Doppler ultrasound measurement of cerebral blood flow before the later transition to NIRS technology [7, 93]. As transfer function analysis operates in the frequency domain, it shares the same vulnerability to missing data as frequency correlation. The use of interpolation and data windows are effective compensatory mechanisms.
Wavelet coherence
The wavelet coherence approach to autoregulation can be conceptualized as an alternative to frequency correlation while also incorporating the element of time, ultimately providing a hybrid of time and frequency correlation. Rather than decomposing a complex waveform into a series of sine waves of varying frequency and amplitude, wavelet transformation breaks this signal into small “wavelets” or brief bursts limited in time and frequency. Hundreds of possible wavelet shapes can be scaled to match the underlying signal. By taking the wavelet transformation of a signal’s autocorrelation function, the wavelet power spectrum can be obtained and is analogous to the power spectrum obtained through Fourier analysis. Coherence and transfer function analysis can be performed using the wavelet transformation of the NIRS and blood pressure signals. The strength of coherence or transfer function gain can then be visualized across frequencies and time and has been implemented using conventional and broadband NIRS in several studies of infants with HIE [94,95,96]. As with other approaches utilizing the frequency domain, wavelet analysis is not robust to missing data. The same set of tools used in other approaches (windowed data, interpolation) are equally useful in this approach.
Challenges to autoregulation methods
All the described methods quantify cerebral autoregulatory function. A significant challenge in the literature is the lack of simultaneous comparison of different methodologies to the same patient population to compare performance characteristics. What limited data exist in adult and animal populations suggest a general equivalence between compared approaches [97,98,99]. There are a number of known challenges with the described approaches [100]. Time correlation, frequency correlation, and cerebrovascular reactivity use thresholds to calculate autoregulatory function and a correlation greater than 0.5 is most used to identify periods of impaired autoregulation. Although recent efforts have been made to derive the ideal threshold based on statistical calculations [9, 75] the optimal value, if it exists remains elusive [101]. Regardless of the numeric value of the threshold, the mere existence of a threshold implies modeling of autoregulation as a binary function—either on or off. While this is mathematically simpler, it is likely an incomplete picture of the underlying biology.
Transfer function and wavelet coherence approach overcome this problem by less reliance on thresholds, instead of providing a continuous scale output. While this is advantageous, implementation of these approaches requires significantly greater technical expertise and the resulting measurement is conceptually more complex, providing a barrier to future clinical adoption.
Conclusion
NIRS is a valuable tool for estimating cerebral oxygenation and for modeling cerebral autoregulation. The availability of specific neonatal probes and the non-invasive nature of commercial NIRS devices make them particularly well suited for use in both term and preterm neonates. When properly implemented, it offers the potential to assess mechanisms underlying brain injury in this population and allows the informed design of novel neuroprotective strategies.
In Table 4, we have listed our recommendations. Although there is no governing organization to issue standards in this field, it is our hope that these recommendations provide a starting point for future neonatal NIRS research. Using a common framework to record, process, and analyze data will allow published results to be readily compared and facilitate future collaborations.
In addition to the harmonization of data capture, additional questions remain. In the data processing context, more research is needed to truly define the best method for handling missing or erroneous data. An active investigation is also needed to identify the best method for quantifying autoregulation in neonates at risk for brain injury. Choice of analytic methods should be driven by the objectives of the study and the nature of the captured data. A large, comparative study of the different approaches is required to fully evaluate the superiority (or equivalence) of any given method. Finally, there is a significant promise on the horizon for emerging NIRS technologies that will offer the ability to examine cerebral oxygenation and blood flow using novel approaches.
References
Marin T, Moore J. Understanding near-infrared spectroscopy. Adv Neonatal Care. 2011;11:382–8.
Taussky P, O’Neal B, Daugherty WP, Luke S, Thorpe D, Pooley RA, et al. Validation of frontal near-infrared spectroscopy as noninvasive bedside monitoring for regional cerebral blood flow in brain-injured patients. Neurosurg Focus. 2012;32:E2.
Calderon-Arnulphi M, Alaraj A, Slavin KV. Near infrared technology in neuroscience: past, present and future. Neurol Res. 2009;31:605–14.
Greisen G, Leung T, Wolf M. Has the time come to use near-infrared spectroscopy as a routine clinical tool in preterm infants undergoing intensive care? Philos Trans A Math Phys Eng Sci. 2011;369:4440–51.
van Bel F, Lemmers P, Naulaers G. Monitoring neonatal regional cerebral oxygen saturation in clinical practice: value and pitfalls. Neonatology. 2008;94:237–44.
Chock VY, Ramamoorthy C, Van, Meurs KP. Cerebral autoregulation in neonates with a hemodynamically significant patent ductus arteriosus. J Pediatr. 2012;160:936–42.
Vesoulis ZA, Liao SM, Trivedi SB, Ters NE, Mathur AM. A novel method for assessing cerebral autoregulation in preterm infants using transfer function analysis. Pediatr Res. 2016;79:453–9.
Lee JK, Kibler KK, Benni PB, Easley RB, Czosnyka M, Smielewski P, et al. Cerebrovascular reactivity measured by near-infrared spectroscopy. Stroke. 2009;40:1820–6.
Soul JS, Hammer PE, Tsuji M, Saul JP, Bassan H, Limperopoulos C, et al. Fluctuating pressure-passivity is common in the cerebral circulation of sick premature infants. Pediatr Res. 2007;61:467–73.
Wong FY, Leung TS, Austin T, Wilkinson M, Meek JH, Wyatt JS, et al. Impaired autoregulation in preterm infants identified by using spatially resolved spectroscopy. Pediatrics. 2008;121:e604–11.
Burns DA, Ciurczak EW, editors. Handbook of near-infrared analysis. 3rd ed. Boca Raton: CRC Press, 2008.
Rolfe P, YABD Wickramasinghe, Thorniley MS, Faris F, Houston R, Kai Z, et al. Fetal and neonatal cerebral oxygen monitoring with NIRS: theory and practice. Early Hum Dev. 1992;29:269–73.
Bass M, Mahajan VN. Handbook of optics. Volume I. New York: McGraw-Hill, 2010 http://www.dawsonera.com/depp/reader/protected/external/AbstractView/S9780071629256. Accessed 10 Feb 2020.
Suzuki S, Takasaki S, Ozaki T, Kobayashi Y. Tissue oxygenation monitor using NIR spatially resolved spectroscopy. In: Chance B, Alfano RR, Tromberg BJ editors. San Jose, CA, 1999. p. 582–92.
Watzman HM, Kurth CD, Montenegro LM, Rome J, Steven JM, Nicolson SC. Arterial and venous contributions to near-infrared cerebral oximetry. Anesthesiology. 2000;93:947–53.
Wong FY, Alexiou T, Samarasinghe T, Brodecky V, Walker AM. Cerebral arterial and venous contributions to tissue oxygenation index measured using spatially resolved spectroscopy in newborn lambs. Anesthesiology. 2010;113:1385–91.
Ikeda K, MacLeod DB, Grocott HP, Moretti EW, Ames W, Vacchiano C. The accuracy of a near-infrared spectroscopy cerebral oximetry device and its potential value for estimating jugular venous oxygen saturation. Anesth Analg. 2014;119:1381–92.
Grubhofer G, Tonninger W, Keznickl P, Skyllouriotis P, Ehrlich M, Hiesmayr M, et al. A comparison of the monitors INVOS 3100 and NIRO 500 in detecting changes in cerebral oxygenation. Acta Anaesthesiol Scand. 1999;43:470–5.
Cho H, Nemoto EM, Sanders M, Fernandez K, Yonas H. Comparison of two commercially available near-infrared spectroscopy instruments for cerebral oximetry. Technical note. J Neurosurg. 2000;93:351–4.
Dix LML, van Bel F, Baerts W, Lemmers PMA. Comparing near-infrared spectroscopy devices and their sensors for monitoring regional cerebral oxygen saturation in the neonate. Pediatr Res. 2013;74:557–63.
Hessel TW, Hyttel-Sorensen S, Greisen G. Cerebral oxygenation after birth—a comparison of INVOS® and FORE-SIGHTTM near-infrared spectroscopy oximeters. Acta Paediatr. 2014;103:488–93.
Kleiser S, Nasseri N, Andresen B, Greisen G, Wolf M. Comparison of tissue oximeters on a liquid phantom with adjustable optical properties. Biomed Opt Express. 2016;7:2973–92.
Kleiser S, Ostojic D, Andresen B, Nasseri N, Isler H, Scholkmann F, et al. Comparison of tissue oximeters on a liquid phantom with adjustable optical properties: an extension. Biomed Opt Express. 2018;9:86–101.
Hyttel-Sorensen S, Kleiser S, Wolf M, Greisen G. Calibration of a prototype NIRS oximeter against two commercial devices on a blood-lipid phantom. Biomed Opt Express. 2013;4:1662–72.
Durduran T, Yodh AG. Diffuse correlation spectroscopy for non-invasive, micro-vascular cerebral blood flow measurement. NeuroImage. 2014;85:51–63.
Giovannella M, Andresen B, Andersen JB, El-Mahdaoui S, Contini D, Spinelli L, et al. Validation of diffuse correlation spectroscopy against 15 O-water PET for regional cerebral blood flow measurement in neonatal piglets. J Cereb Blood Flow Metab. 2020;40:2055–65.
Buckley EM, Parthasarathy AB, Grant PE, Yodh AG, Franceschini MA. Diffuse correlation spectroscopy for measurement of cerebral blood flow: future prospects. Neurophoton. 2014;1:011009.
Durduran T, Zhou C, Buckley EM, Kim MN, Yu G, Choe R, et al. Optical measurement of cerebral hemodynamics and oxygen metabolism in neonates with congenital heart defects. J Biomed Opt. 2010;15:037004.
Giovannella M. BabyLux device: a diffuse optical system integrating diffuse correlation spectroscopy and time-resolved near-infrared spectroscopy for the neuromonitoring of the premature newborn brain. Neurophoton. 2019;6:1.
Lee CW, Cooper RJ, Austin T. Diffuse optical tomography to investigate the newborn brain. Pediatr Res. 2017;82:376–86.
Liao SM, Gregg NM, White BR, Zeff BW, Bjerkaas KA, Inder TE, et al. Neonatal hemodynamic response to visual cortex activity: high-density near-infrared spectroscopy study. J Biomed Opt. 2010;15:026010.
Dempsey LA, Cooper RJ, Powell S, et al. Whole-head functional brain imaging of neonates at cot-side using time-resolved diffuse optical tomography. Diffuse optical imaging V. OSA: Munich, 2015. p. 953818.
Singh H, Cooper RJ, Wai Lee C, Dempsey L, Edwards A, Brigadoi S, et al. Mapping cortical haemodynamics during neonatal seizures using diffuse optical tomography: a case study. Neuroimage Clin. 2014;5:256–65.
Hintz SR, Cheong W-F, Van Houten JP, Stevenson DK, Benaron DA. Bedside imaging of intracranial hemorrhage in the neonate using light: comparison with ultrasound, computed tomography, and magnetic resonance imaging. Pediatr Res. 1999;45:54–9.
Bale G, Mitra S, Meek J, Robertson N, Tachtsidis I. A new broadband near-infrared spectroscopy system for in-vivo measurements of cerebral cytochrome-c-oxidase changes in neonatal brain injury. Biomed Opt Express. 2014;5:3450.
Mitra S, Bale G, Meek J, Tachtsidis I, Robertson NJ. Cerebral near infrared spectroscopy monitoring in term infants with hypoxic ischemic encephalopathy—a systematic review. Front Neurol. 2020;11:393.
Bale G, Rajaram A, Kewin M, Morrison L, Bainbridge A, Diop M, et al. Broadband NIRS cerebral cytochrome-c-oxidase response to anoxia before and after hypoxic-ischaemic injury in piglets. Adv Exp Med Biol. 2018;1072:151–6.
Bale G, Mitra S, de Roever I, Chan M, Caicedo-Dorado A, Meek J, et al. Interrelationship between broadband NIRS measurements of cerebral cytochrome C oxidase and systemic changes indicates injury severity in neonatal encephalopathy. Adv Exp Med Biol. 2016;923:181–6.
Wijbenga RG, Lemmers PMA, van Bel F. Cerebral oxygenation during the first days of life in preterm and term neonates: differences between different brain regions. Pediatr Res. 2011;70:389–94.
Alderliesten T, Dix L, Baerts W, Caicedo Dorado A, van Huffel S, Naulaers G et al. Reference values of regional cerebral oxygen saturation during the first 3 days of life in preterm neonates. Pediatr Res. 2015. https://doi.org/10.1038/pr.2015.186.
Hyttel-Sorensen S, Pellicer A, Alderliesten T, Austin T, van Bel F, Benders M, et al. Cerebral near infrared spectroscopy oximetry in extremely preterm infants: phase II randomised clinical trial. BMJ. 2015;350:g7635.
Citerio G, Park S, Schmidt JM, Moberg R, Suarez JI, Le Roux PD, et al. Data collection and interpretation. Neurocrit Care. 2015;22:360–8.
International Organization for Standardization. ISO 8601-1:2019 date and time—representations for information interchange—part 1: basic rules. 1st ed. 2019. https://www.iso.org/standard/70907.html. Accessed 28 Aug 2020.
Institute of Electrical and Electronics Engineers, Inc, The Open Group. 1003.1 Standard for information technology—portable operating system interface (POSIX) rationale (Informative). IEEE 2001.
Kayaalp M. Modes of de-identification. AMIA Annu Symp Proc. 2017;2017:1044–50.
Garfinkel SL. De-identification of personal information. National Institute of Standards and Technology, 2015.
Singh K, Upadhyaya S. Outlier detection: applications and techniques. Int J Computer Sci Issues. 2012;9:307.
Hegyi T, Carbone MT, Anwar M, Ostfeld B, Hiatt M, Koons A, et al. Blood pressure ranges in premature infants. I. The first hours of life. J Pediatr. 1994;124:627–33.
Zubrow AB, Hulman S, Kushner H, Falkner B. Determinants of blood pressure in infants admitted to neonatal intensive care units: a prospective multicenter study. Philadelphia neonatal blood pressure study group. J Perinatol. 1995;15:470–9.
Batton B, Li L, Newman NS, Das A, Watterberg KL, Yoder BA, et al. Evolving blood pressure dynamics for extremely preterm infants. J Perinatol. 2014;34:301–5.
Martin RJ, Fanaroff AA, Walsh MC. Fanaroff and Martin’s neonatal-perinatal medicine: diseases of the fetus and infant. Philadelphia: Saunders/Elsevier, 2011.
Virtanen J, Noponen T, Kotilahti K, Virtanen J, Ilmoniemi RJ. Accelerometer-based method for correcting signal baseline changes caused by motion artifacts in medical near-infrared spectroscopy. J Biomed Opt. 2011;16:087005.
Siddiquee MR, Marquez JS, Atri R, Ramon R, Perry Mayrand R, Bai O. Movement artefact removal from NIRS signal using multi-channel IMU data. BioMed Eng OnLine. 2018;17:120.
Zhang Y, Brooks DH, Franceschini MA, Boas DA. Eigenvector-based spatial filtering for reduction of physiological interference in diffuse optical imaging. J Biomed Opt. 2005;10:011014.
Yücel MA, Selb J, Cooper RJ, Boas DA. Targeted principle component analysis: a new motion artifact correction approach for near-infrared spectroscopy. J Innov Opt Health Sci. 2014;07:1350066.
Molavi B, Dumont GA. Wavelet-based motion artifact removal for functional near-infrared spectroscopy. Physiol Meas. 2012;33:259–70.
Izzetoglu M, Chitrapu P, Bunce S, Onaral B. Motion artifact cancellation in NIR spectroscopy using discrete Kalman filtering. BioMed Eng OnLine. 2010;9:16.
Ayaz H, Izzetoglu M, Shewokis PA, Onaral B. Sliding-window motion artifact rejection for functional near-infrared spectroscopy. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:6567–70.
Fekete T, Rubin D, Carlson JM, Mujica-Parodi LR. The NIRS analysis package: noise reduction and statistical inference. PLoS ONE. 2011;6:e24322.
Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cyber. 1979;9:62–6.
Blasi A, Phillips D, Lloyd-Fox S, Koh PH, Elwell CE. Automatic detection of motion artifacts in infant functional optical topography studies. In: Takahashi E, Bruley DF editors. Oxygen transport to tissue XXXI. Boston, MA: Springer US, 2010. p. 279–84.
Brigadoi S, Ceccherini L, Cutini S, Scarpa F, Scatturin P, Selb J, et al. Motion artifacts in functional near-infrared spectroscopy: a comparison of motion correction techniques applied to real cognitive data. NeuroImage. 2014;85:181–91.
Cooper RJ, Selb J, Gagnon L, Phillip D, Schytz HW, Iversen HK et al. A systematic comparison of motion artifact correction techniques for functional near-infrared spectroscopy. Front Neurosci. 2012; 6. https://doi.org/10.3389/fnins.2012.00147.
Robertson FC, Douglas TS, Meintjes EM. Motion artifact removal for functional near infrared spectroscopy: a comparison of methods. IEEE Trans Biomed Eng. 2010;57:1377–87.
Woo WL, Koh BHD, Gao B, Nwoye EO, Wei B, Dlay SS. Early warning of health condition and visual analytics for multivariable vital signs. In: Proceedings of the 2020 international conference on computing, networks and internet of things. Sanya China: ACM, 2020. p. 206–11.
Lomb NR. Least-squares frequency analysis of unequally spaced data. Astrophys Space Sci. 1976;39:447–62.
Jahani S, Setarehdan SK, Boas DA, Yücel MA. Motion artifact detection and correction in functional near-infrared spectroscopy: a new hybrid method based on spline interpolation method and Savitzky–Golay filtering. Neurophotonics. 2018;5:015003.
Scholkmann F, Spichtig S, Muehlemann T, Wolf M. How to detect and reduce movement artifacts in near-infrared imaging using moving standard deviation and spline interpolation. Physiol Meas. 2010;31:649–62.
Hemphill JC, Barton CW, Morabito D, Manley GT. Influence of data resolution and interpolation method on assessment of secondary brain insults in neurocritical care. Physiol Meas. 2005;26:373–86.
Moore L, Hanley JA, Lavoie A, Turgeon A. Evaluating the validity of multiple imputation for missing physiological data in the national trauma data bank. J Emerg Trauma Shock. 2009;2:73–9.
Tan FES, Jolani S, Verbeek H. Guidelines for multiple imputations in repeated measurements with time-dependent covariates: a case study. J Clin Epidemiol. 2018;102:107–14.
Vaseghi SV. Advanced digital signal processing and noise reduction. 4th ed. Chichester, UK: J. Wiley & Sons, 2008.
Paiement A, Mirmehdi M, Xianghua Xie, Hamilton MCK. Integrated segmentation and interpolation of sparse data. IEEE Trans Image Process. 2014;23:110–25.
Massaro AN, Govindan RB, Vezina G, Chang T, Andescavage NN, Wang Y, et al. Impaired cerebral autoregulation and brain injury in newborns with hypoxic-ischemic encephalopathy treated with hypothermia. J Neurophysiol. 2015;114:818–24.
Govindan RB, Massaro AN, Andescavage NN, Chang T, du Plessis A. Cerebral pressure passivity in newborns with encephalopathy undergoing therapeutic hypothermia. Front Hum Neurosci. 2014; 8. https://doi.org/10.3389/fnhum.2014.00266.
Alosh H, Ramirez A, Mink R. The correlation between brain near-infrared spectroscopy and cerebral blood flow in piglets with intracranial hypertension. J Appl Physiol. 2016;121:255–60.
Zernikow B, Michel E, Jorch G. Cerebral autoregulation is a nonlinear type control system. Pediatr Res. 1994;35:275–5.
Czosnyka M, Smielewski P, Kirkpatrick P, Menon DK, Pickard JD. Monitoring of cerebral autoregulation in head-injured patients. Stroke. 1996;27:1829–34.
Czosnyka M, Smielewski P, Kirkpatrick P, Laing RJ, Menon D, Pickard JD. Continuous assessment of the cerebral vasomotor reactivity in head injury. Neurosurgery. 1997;41:11–19.
Brady KM, Lee JK, Kibler KK, Smielewski P, Czosnyka M, Easley RB, et al. Continuous time-domain analysis of cerebrovascular autoregulation using near-infrared spectroscopy. Stroke. 2007;38:2818–25.
Rhee CJ, Fraser Iii CD, Kibler K, Easley RB, Andropoulos DB, Czosnyka M, et al. The ontogeny of cerebrovascular pressure autoregulation in premature infants. J Perinatol. 2014;34:926–31.
Eriksen VR, Hahn GH, Greisen G. Dopamine therapy is associated with impaired cerebral autoregulation in preterm infants. Acta Paediatr. 2014;103:1221–6.
Gilmore MM, Stone BS, Shepard JA, Czosnyka M, Easley RB, Brady KM. Relationship between cerebrovascular dysautoregulation and arterial blood pressure in the premature infant. J Perinatol. 2011;31:722–9.
Howlett JA, Northington FJ, Gilmore MM, Tekes A, Huisman TAGM, Parkinson C. et al. Cerebrovascular autoregulation and neurologic injury in neonatal hypoxic-ischemic encephalopathy. Pediatr Res. 2013;74:525–35.
Burton VJ, Gerner G, Cristofalo E, Chung S, Jennings JM, Parkinson C, et al. A pilot cohort study of cerebral autoregulation and 2-year neurodevelopmental outcomes in neonates with hypoxic-ischemic encephalopathy who received therapeutic hypothermia. BMC Neurol. 2015;15:209.
Welch P. The use of fast Fourier transform for the estimation of power spectra: a method based on time averaging over short, modified periodograms. IEEE Trans Audio Electroacoust. 1967;15:70–3.
Smet DD, Vanderhaegen J, Naulaers G, Huffel SV. New measurements for assessment of impaired cerebral autoregulation using near-infrared spectroscopy. In: Liss P, Hansell P, Bruley DF, Harrison DK editors. Oxygen transport to tissue XXX. Boston, MA: Springer US, 2009, p. 273–78.
Caicedo A, De Smet D, Vanderhaegen J, Naulaers G, Wolf M, Lemmers P et al. Impaired cerebral autoregulation using near-infrared spectroscopy and its relation to clinical outcomes in premature infants. In: LaManna JC, Puchowicz MA, Xu K, Harrison DK, Bruley DF editors. Oxygen transport to tissue XXXII. Springer US: Boston, MA, 2011. p. 233–9.
Grossbard NJ, Dewan EM. Methods for estimating the autocorrelation and power spectral density functions when there are many missing data values. In: Proceedings of the fifth ASSP workshop on spectrum estimation and modeling. IEEE: Rochester, NY, USA, 1990. p. 30–4.
Lee TCM, Zhengyuan Zhu. Nonparametric spectral density estimation with missing observations. In: Proceedings of the 2009 IEEE international conference on acoustics, speech and signal processing. IEEE: Taipei, Taiwan, 2009. p. 3041–44.
JAHR Claassen, Meel-van den Abeelen ASS, Simpson DM, Panerai RB. International cerebral autoregulation research network (CARNet). Transfer function analysis of dynamic cerebral autoregulation: a white paper from the international cerebral autoregulation research network. J Cereb Blood Flow Metab. 2016;36:665–80.
Panerai RB, Hudson V, Fan L, Mahony P, Yeoman PM, Hope T, et al. Assessment of dynamic cerebral autoregulation based on spontaneous fluctuations in arterial blood pressure and intracranial pressure. Physiol Meas. 2002;23:59–72.
Vesoulis ZA, Liao SM, Mathur AM. Delayed cord clamping is associated with improved dynamic cerebral autoregulation and decreased incidence of intraventricular hemorrhage in preterm infants. J Appl Physiol. 2019;127:103–10.
Tian F, Tarumi T, Liu H, Zhang R, Chalak L. Wavelet coherence analysis of dynamic cerebral autoregulation in neonatal hypoxic–ischemic encephalopathy. NeuroImage: Clin. 2016;11:124–32.
Chalak LF, Zhang R. New wavelet neurovascular bundle for bedside evaluation of cerebral autoregulation and neurovascular coupling in newborns with hypoxic-ischemic encephalopathy. Dev Neurosci. 2017;39:89–96.
Mitra S, Bale G, Highton D, Gunny R, Uria-Avellanal C, Bainbridge A et al. Pressure passivity of cerebral mitochondrial metabolism is associated with poor outcome following perinatal hypoxic ischemic brain injury. J Cereb Blood Flow Metab. 2017; 0271678X1773363.
Brady KM, Lee JK, Kibler KK, Easley RB, Koehler RC, Shaffner DH. Continuous measurement of autoregulation by spontaneous fluctuations in cerebral perfusion pressure: comparison of 3 methods. Stroke. 2008;39:2531–27.
Liu X, Czosnyka M, Donnelly J, Budohoski KP, Varsos GV, Nasr N, et al. Comparison of frequency and time domain methods of assessment of cerebral autoregulation in traumatic brain injury. J Cereb Blood Flow Metab. 2015;35:248–56.
Liu X, Czosnyka M, Donnelly J, Cardim D, Cabeleira M, Hutchinson PJ, et al. Wavelet pressure reactivity index: a validation study: wavelet pressure reactivity index. J Physiol. 2018;596:2797–809.
Czosnyka M, Brady K, Reinhard M, Smielewski P, Steiner LA. Monitoring of cerebrovascular autoregulation: facts, myths, and missing links. Neurocrit Care. 2009;10:373–86.
Kooi EMW, Verhagen EA, Elting JWJ, Czosnyka M, Austin T, Wong FY, et al. Measuring cerebrovascular autoregulation in preterm infants using near-infrared spectroscopy: an overview of the literature. Expert Rev Neurother. 2017;17:801–18.
Funding
NIH Grant K23 NS111086 (ZAV). NIH Grant 1R01HL12216701A1 (VYC).
Author information
Authors and Affiliations
Contributions
The conception of an article (ZAV, JPM, and VYC); initial drafting of manuscript (ZAV); creation of figures and tables (ZAV, VYC); critical revision and approval of submitted final manuscript (ZAV, JPM, and VYC).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vesoulis, Z.A., Mintzer, J.P. & Chock, V.Y. Neonatal NIRS monitoring: recommendations for data capture and review of analytics. J Perinatol 41, 675–688 (2021). https://doi.org/10.1038/s41372-021-00946-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41372-021-00946-6
This article is cited by
-
Neonatal somatic oxygenation and perfusion assessment using near-infrared spectroscopy
Pediatric Research (2024)
-
Aeration strategy at birth does not impact carotid haemodynamics in preterm lambs
Pediatric Research (2023)
-
Preterm infants variability in cerebral near-infrared spectroscopy measurements in the first 72-h after birth
Pediatric Research (2023)
-
Advanced Neuromonitoring Modalities on the Horizon: Detection and Management of Acute Brain Injury in Children
Neurocritical Care (2023)
-
Therapeutic Hypothermia in a Tertiary Reference Center of Rio de Janeiro
Current Pediatrics Reports (2023)