Abstract
Ginsenoside Rg1 (Rg1), a saponin extracted from Panax ginseng, has been well documented to be effective against ischemic/reperfusion (I/R) neuronal injury. However, the underlying mechanisms remain obscure. In the present study, we investigated the roles of Nrf2 and miR-144 in the protective effects of Rg1 against I/R-induced neuronal injury. In OGD/R-treated PC12 cells, Rg1 (0.01–1 μmol/L) dose-dependently attenuated the cell injury accompanied by prolonging nuclear accumulation of Nrf2, enhancing the transcriptional activity of Nrf2, as well as promoting the expression of ARE-target genes. The activation of the Nrf2/ARE pathway by Rg1 was independent of disassociation with Keap1, but resulted from post-translational regulations. Knockdown of Nrf2 abolished all the protective changes of Rg1 in OGD/R-treated PC12 cells. Furthermore, Rg1 treatment significantly decreased the expression of miR-144, which downregulated Nrf2 production by targeting its 3’-untranlated region after OGD/R. Knockdown of Nrf2 had no effect on the expression of miR-144, suggesting that miR-144 was an upstream regulator of Nrf2. We revealed that there was a direct binding between Nrf2 and miR-144 in PC12 cells. Application of anti-miR-144 occluded the activation of the Nrf2/ARE pathway by Rg1 in OGD/R-treated PC12 cells. In tMCAO rats, administration of Rg1 (20 mg/kg) significantly alleviated ischemic injury, and activated Nrf2/ARE pathway. The protective effects of Rg1 were abolished by injecting of AAV-HIF-miR-144-shRNA into the predicted ischemic penumbra. In conclusion, our results demonstrate that Rg1 alleviates oxidative stress after I/R through inhibiting miR-144 activity and subsequently promoting the Nrf2/ARE pathway at the post-translational level.
Similar content being viewed by others
Introduction
Panax Ginseng has been used for thousands of years in traditional Chinese medicine as a tonic to improve stamina and vitality [1, 2]. Ginsenoside Rg1 (Rg1), a purified saponin isolated from ginseng, is considered one of the most potent antistroke candidates by many recent studies [3,4,5]. A systematic review and meta-analysis revealed a marked efficacy of Rg1 in experimental acute ischemic stroke, as manifested by its ability to reduce infarct volume and improve neurological score [6, 7]. However, the underlying mechanism is still unclear.
Evidence has shown that oxidative stress is closely related to the pathological progression of ischemia/reperfusion (I/R) injury [8,9,10,11], which arises from excessive production or deficient degradation of reactive oxygen species (ROS) [12]. Under physiological conditions, there is a balance between ROS generation and ROS elimination by scavengers. I/R disrupts this balance from the initial stage of I/R, as evidenced by increased ROS production, consumption of antioxidants, and finally, the induction of oxidative damage to proteins, lipids, DNA, and RNA [13,14,15]. ROS scavengers, including edaravone and salvianolic acids, have been shown to be beneficial for the treatment of ischemic stroke in clinical trials [15,16,17].
In addition to scavenging ROS directly, the activation of Nrf2 and the expression of its downstream genes may have protective effects against I/R damage, as validated by emerging evidence [18, 19]. As one of the master regulators for defending against oxidative stress, Nrf2 binds to the antioxidant response element (ARE) in the nucleus and promotes a series of antioxidation gene expression [20]. The level of Nrf2 showed an inverted U shape after ischemic stroke, characterized by a robust increase from 2 h of OGD, a peak at 8 h, and then a decrease below baseline at ~ 24 h in transient MCAO mice [21]. Loss of Nrf2 exacerbated cerebral infarction and neurologic deficits in both transient and permanent middle cerebral artery occlusion (tMCAO and pMCAO, respectively) models, as revealed by a reduction in oxidative stress tolerance, increased TLR4 expression, NF-κB activation, breakdown of the blood–brain barrier, and other supporting evidence [22,23,24]. Activation of Nrf2 and its target genes may protect the brain against I/R damage [25, 26]. Hence, Nrf2 is considered a promising therapeutic target for defending against the oxidative stress generated during a stroke.
In addition to Keap1, the Nrf2/ARE pathway is regulated by many other factors, such as DNA methylation, histone acetylation and microRNA (miRNA) regulation [27]. These miRNAs are a group of endogenous small noncoding RNAs that negatively control gene expression by binding to their target sequences at the 3′-UTR of mRNAs [28]. Until now, it had been proven that miR-144, miR-153, and miR-27 directly regulate Nrf2 production in neurodegenerative diseases [29, 30]. Among them, miR-144 and miR-153 showed related pathological changes with ischemic impairment. An increase in miR-153 was also found in hippocampal neurons subjected to oxygen glucose deprivation/reperfusion (OGD/R) treatment [31]. However, application of Rg1 showed no effect on the level of miR-153 (data not shown). The absence of miR-144 impaired ischemic preconditioning-induced cardio-protection [32], and it was also involved in the pathogenesis of Alzheimer’s disease (the most common neurodegenerative disease) [33]. This suggested that miR-144 may be involved in the pathogenesis of ischemic injury to the central nervous system. Therefore, in this research, miR-144 was selected as the miRNA of interest.
In the present study, we demonstrated that the Nrf2/ARE pathway was activated by Rg1 treatment both in vitro and in vivo. Inhibition of Nrf2 significantly reduced Rg1’s effect. Interestingly rather than Keap1, one of the miRNAs, miR-144, interacted with Nrf2 and was involved in this regulating process. Furthermore, we found that the expression of miRNA-144 was independent from the activation of Nrf2, indicating that miR-144 is an upstream regulator of Nrf2 in I/R injury. Inhibition of miR-144 erased the anti-I/R effect of Rg1, as well as the activation of the Nrf2/ARE pathway in vivo. All of these results suggest that oxidative stress after I/R was alleviated by Rg1 by promoting the antioxidative stress defenses of the Nrf2/ARE pathway at the post-translational level by inhibiting miR-144 activity. This provides a new drug-development target for Rg1 application for the potential treatment of ischemic stroke.
Materials and methods
Reagents
Rg1 (HPLC 98%) was provided by Jecui Healthy Co., Ltd. (Yunnan, China), and its chemical structure is shown in Fig. 1a. Dulbecco's Modified Eagle's Medium (DMEM) and fetal bovine serum were purchased from Gibco (Grand Island, NY, USA). β-Actin primary antibody, ascorbic acid, momotetrazolium (MTT), and dimethylsulfoxide (DMSO) were purchased from Sigma (St. Louis, MO, USA). The test kits for measurement of lactic dehydrogenase (LDH) were purchased from Nanjing Institute of Jiancheng Bioengineering (Nanjing, China). Primary antibodies against Nrf2, NQO-1, HO-1, glutamate cysteine ligase modifier subunit (GCLM), glutamate cysteine ligase catalytic subunit (GCLC), and Lamin A were purchased from Abcam (Cambridge, UK). All other chemicals and solvents used were of analytical grade unless otherwise specified.
Rg1 protects against OGD/R-induced neurotoxicity in PC12 cells. a The chemical structure of Rg1. MTT assay b and LDH release determination c were employed to investigate the protective effect of Rg1 against OGD/R. The level of intracellular ROS was also revealed by DCFH-DA staining d. All data were presented as mean ± SD of triplicate independent experiments. One-way ANOVA analysis followed by Newman–Keuls test (## P < 0.01, ### P< 0.001 vs. control group, * P< 0.05, ** P< 0.01 vs. model group)
PC12 cell culture and OGD/R
Rat pheochromocytoma PC12 cells were obtained from Peking Union Medical College. The OGD/R was performed as previously described [34]. In brief, PC12 cells were cultured in DMEM with 10% fetal bovine serum, 5% embryonic stem, 100 U/mL penicillin, and 100 U/mL streptomycin at 37 °C under a humidified atmosphere containing 5% CO2. For OGD, cells were cultured in glucose-free Earle’s balanced salt solution supplemented with 20 mM Na2S2O4 for 20 min. After that, they were incubated with conditioned DMEM with or without Rg1 at different concentrations (0.01, 0.10, 1.00, and 10.00 µM) for the indicated time.
Cell viability determination
Cell viability was measured with an MTT assay. In brief, PC12 cells were seeded in 96-well plates (5 × 103 cells/well) and incubated for 24 h. The cells then underwent 20 min of OGR, followed by incubation with conditioned DMEM with or without Rg1 at different concentrations (0.01, 0.10, 1.00, and 10.00 µM) for 24 h. After that, they were incubated at 37 °C for 4 h in 0.5 mg/mL MTT solution, and the medium was carefully removed. Then, 100 μL of DMSO was added to each well to dissolve the violet formazan crystals. The absorbance at 570 nm was measured. All values were normalized to the control group.
OGD/R-induced neurotoxicity was also quantified by measuring LDH release from damaged cells according to the manufacture’s instruction (Nanjing Institute of Jiancheng Bioengineering). In brief, 150 μL of LDH reaction reagent was added to 50 μL medium taken from the cell culture wells. The absorbance was measured at 490 nm by a spectrophotometer plate reader. Data were expressed in the percentage of cell death that was calculated according to the manufacturer’s instruction.
ROS determination
Intracellular ROS level was detected using the 20,70-dichlorodihydrofluorescein diacetate (DCFH-DA) fluorescence assay. In brief, cells were seeded in 96-well plates, overnight at a density of 7 × 103 per well. Rg1 (0.01, 0.10, 1.00, and 10.00 µM) was added into culture for 24 h followed by OGD. After that, the cells were incubated in serum-free medium containing 25 μM DCFH-DA at 37 °C for 30 min. ROS levels were measured by the fluorescence intensity of DCF at 525 nm after excitation at 488 nm on a fluorescence plate reader (Thermo Scientific Varioskan Flash).
Nuclei and cytosol extraction from culture cells
Nuclear and cytosolic protein fractionation was prepared from PC12 cells or brain samples using the nuclear-cytosol extraction kit (Applygen Technologies Inc, Beijing, China) according to the manufacturer’s instructions. Cells were washed with phosphate-buffered saline (PBS), followed by detachment with trypsinization and centrifugation for 5 min at 800 × g to collect cells. Then, 250 μL Cytosol Extraction Buffer A was added to the cells and vortexed for 30 s; the samples were then incubated on ice for 10 min. Next, 30 μL of Cytosol Extraction Buffer B was added to the mixture and vortexed for 10 s, followed by incubation on ice for 1 min. The tube was centrifuged for 5 min at 1000 × g at 4 °C. The pellet and supernatant were treated separately. Brain samples were lysed by adding 10 times volume/quantity of cytosol extraction buffer A, and the subsequent protocol was the same as previously described [35].
Western blot analysis
Proteins were separated depending on their size by sodium dodecyl sulfate polyacrylamide gel electrophoresis on 15% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore, USA). The membranes were blocked with 3% bovine serum albumin and incubated with primary antibodies and then horseradish peroxidase-conjugated secondary antibodies. Proteins were then detected with an enhanced chemiluminescence plus detection system (PPLYGEN, China). The signal protein bands were scanned and analyzed with Gel-Pro Analyzer software (Media Cybernetics).
RNA extraction and real-time PCR
Total cellular RNA was extracted using Trizol reagent. RNA was reverse transcribed to cDNA using a Quantitect reverse transcription kit according to the manufacturer’s instructions (Qiagen). For real-time PCR analysis of HO-1, NQO-1, GCLC and GCLM, the specific primers are listed below (5'-3'): rHO-1: forward (AGCGAAACAAGCAGAACCCA) and reverse (ACCTCGTGGAGACGCTTTAC); rGCLM: forward (CACAACTCAGGGGCCTTGTA) and reverse: (AAACCACCACATTCACGCCT); rGCLC: forward (GAGCGAGATGCCGTCTTACA) and reverse (AACACGCCTTCCTTCCCATT); rNQO-1: forward (GCCATCATTTGGGCAAGTCC) and reverse (TCCTTGTGGAACAAAGGCGA); rGAPDH: forward (CCGTATTCAGCATTCTATGCTCTC), and reverse (TGGATACACACTCTGGGGCT). The mRNA levels were analyzed by real-time quantitative RT-PCR using a Bio-Red iCycler system (Bio-Rad, Hercules, CA, USA). miRNA detection by real-time analysis involved reverse transcription using a small RNA-specific stem-loop RT primer. Individual miRNA was detected using Tagman small RNA assay real-time PCR analysis. The results were normalized to small nuclear RNA U6, and the data were expressed as log twofold-changes in respective miRs/U6 mRNA levels.
Dual luciferase reporter assay
Cells were seeded in 96-well plates, incubated for 24 h, and then transfected with various constructs as indicated in the respective figures. Forty-eight hours after transfection, cells were lysed with passive lysis buffer for 20 min. Firefly and Renilla luciferase activities were determined from lysates using dual luciferase assay system (Promega, USA) in a Glomax luminometer.
Small interference RNA and transfection
Nrf2-siRNA or control-siRNA was purchased from Santa Cruz (Santa Cruz Biotechnology). Transfection was performed using Lipofectamine 2000 according to the manufacturer’s protocol. In brief, PC12 cells were transfected with Nrf2-siRNA or control-siRNA for 48 h, followed by treatment with Rg1 (1 μM) for the indicated times.
Animals
Male Sprague-Dawley (SD) rats (6 weeks; 160 to 180 g) were supplied by the Experimental Animal Center of the Chinese Academy of Medical Sciences (Beijing, China). They were housed in a 12-h light/dark scheduled room that is temperature (22 ± 2 °C) and humidity ( < 40%) controlled with free access to food and water. All experimental procedures were performed according to the standards established in the Guide for the Care and Use of Laboratory Animals published by the Institute of Laboratory Animal Resources of the National Research Council (USA) and approved by the local Animal Care and Use Committee of the Peking Union Medical College and the Chinese Academy of Medical Sciences. A computer-generated randomization schedule was used to assign the animals to different groups. The assessments of measuring infarct volume and scoring neurobehavioral outcome were blinded.
Conditional knockout of miR-144 in predicted penumbra zone
According to the report by Ashwal et al. [36, 37], the penumbra zone was mainly located in the middle cortex. To exclude the effects on miR-144 in nonischemic regions, miR-144-shRNA expression was initiated by the HIF1A promoter using pAAV-HIF1A promoter-EGFP-3FLAG-micro30 (miR-144), which was provided by Obio Technology (Shanghai) Corp., Ltd. (China). For viral injections, male SD rats were anesthetized with 10% chloral hydrate (4 mL/kg of body weight, i.p. injection) and injected with 1 μL of AAV virus into the cortex (stereotaxic coordinates: AP, −3.84 mm; ML, + 2.0 mm; DV, − 1.2 mm). The injection sites were examined at the end of all the behavior tests and only data from animals with correct injections were included.
Focal brain ischemia
The transient middle cerebral artery occlusion (tMCAO) model was performed the same as previously described with modifications [38]. In brief, the animal was anesthetized with 10% chloral hydrate (4 mL/kg, ip injection). The tip of a 4–0 nylon monofilament was burned (diameter 0.36 mm) and was inserted into the right common carotid artery. The monofilament was advanced into the origin of the right middle cerebral artery. The thread was removed to allow reperfusion 90 min later, and the animals were returned to their cages.
Neurological function
Longa’s five-point scale was used to evaluate the neurological score [39]. The animals without symptoms of neural damage or death after the surgery were excluded, and other rats were recruited.
Infarct analysis
At the indicated time points (24 h after the tMCAO), brains were removed under anesthesia and cut into 2-mm slices, with six slices per brain. The slices were stained with a 1% solution of 2,3,5-triphenyltetrazolium chloride in PBS at 37 °C for 30 min and fixed in 4% phosphate-buffered formalin. Images of the samples were captured with an electronic scanner and computer. The areas of ipsilateral and contralateral corresponding structures and infarct area were calculated with ImageJ. The infarct area was corrected to account for edema postischemia and shrinkage owing to fixing. The infarct area was corrected using the following formula for both edema and shrinkage: corrected infarct area = measured infarct area + area of contralateral corresponding structure − area of ipsilateral corresponding structure [16]. The infarct volume in each brain was calculated by summing the brain slice thickness (2 mm) and total of infarct area in all sample slices.
ROS production measurement by dihydroethidium (DHE) staining
ROS in brain slices were revealed by DHE. In brief, a dewaxed and dehydrated paraffin section (4 µm) was exposed to DHE (100 μM, Molecular Probes, Sigma, D7008) on the sample surface away from light for 3 h at 37 °C, followed by three washes with PBS with Tween 20 (PBST). Next, the sections were incubated with Hoechst (1:1000) for 15 min at 37 °C. The slides were rinsed three times with PBST, mounted with 90% glycerin and coverslipped. The tissue sections were then visualized, and fluorescence was detected by a laser scanning confocal microscope (Leica TCSSP2, Solms, Germany). The entire quantifying procedure was performed in a blinded manner.
Immunofluorescence staining for DNA oxidative products
DNA oxidative products in brain slices were revealed using immunofluorescence with anti-8-hydroxy-2’-deoxyguanosine (8-OHDG). In brief, dewaxed and dehydrated paraffin sections were incubated for antigen retrieval for 10 min. The slices were then washed three times with PBS at room temperature, 5 min each time. Next, they were soaked in 0.5% Triton X-100 for 10 min and then washed three times with PBS, 5 min each time. The sections were blocked with goat serum and incubated with primary antibodies of 8-OHDG at 4 °C overnight. Subsequently, the sections were washed with PBST three times and incubated for 2 h at room temperature away from light with secondary fluorescence antibodies. All images were captured under a laser scanning confocal microscope (Leica TCSSP2, Solms, Germany). The entire quantifying procedure was performed in a blinded manner.
Statistical analysis
All values are presented as the mean ± SD. Statistical significance between multiple groups was calculated with one-way ANOVA followed by Newman–Keuls post hoc test. Student’s t test was used for testing between two groups. P < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS13.0 software (Chicago, IL, USA)
Results
Rg1 protects against OGD/R-induced neurotoxicity in PC12 cells
To investigate the effect of Rg1 on OGD/R-induced neurotoxicity, PC12 cells were treated with or without Rg1 (structure shown in Fig. 1a) at the indicated concentrations. Neural viability and LDH release were tested by MTT and colorimetry, respectively. The results showed that Rg1 improved PC12 cell viability (Fig. 1b: P < 0.05 vs. model group at 0.01, 0.10, and 1.00 μM) and decreased LDH release (Fig. 1c: P < 0.01 vs model group at 0.10 and 1.00 μM) significantly. However, when the concentration reached 10.00 μM, the protective effect showed a remarkable decreasing tendency. As shown in Fig. 1d, the level of intracellular ROS, determined by DCFH-DA, was significantly increased by OGD/R and was attenuated by Rg1 treatment at the concentration of 0.10 and 1.00 μM (P < 0.05, P < 0.01 vs model group), which is consistent with LDH release. These results suggest that the attenuation of neural viability and LDH release may be related to the changes of ROS level. Moreover, we found that 1.00 μM of Rg1 produced a more favorable effect than any other concentrations in this study.
Rg1 promotes Nrf2 nuclear accumulation without change in Keap1
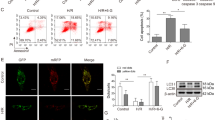
Next, we investigated whether Nrf2 is involved in the anti-OGD/R effect of Rg1 in neurons. PC12 cells were treated with Rg1 or vehicle for different lengths of time under OGD/R, and the change in Nrf2 was evaluated in the nucleus and cytoplasm. As shown in Fig. 2a, b, the nuclear accumulation of Nrf2 significantly increased in the vehicle group from 2 h of OGD, reached its peak at 4 h, and then decreased below baseline at 12 h. Rg1 treatment enhanced the nuclear accumulation of Nrf2, as manifested by prolonged peaking time to 8 h (P < 0.05 vs OGD/R + vehicle group at 8 h) and slowing down the decreasing tendency significantly (P < 0.01 vs OGD/R + vehicle group at 12 h; P < 0.001 vs OGD/R + vehicle group at 24 h). Keap1 is known to be the major endogenous negative regulator for Nrf2. It is responsible for keeping Nrf2 in the cytoplasm and degradation of Nrf2 by binding to the ubiquitin proteasome system [14].
Rg1 promotes Nrf2 nuclear accumulation without change in Keap1. a Representative Western blot images of nuclear Nrf2 accumulation with different duration of OGD/R. Lamin A was used as an internal control. b Quantitative analysis of Nrf2 level in a (Normalized to 0 h for each group). *P < 0.05, ** P< 0.01, *** P< 0.001 vs. OGD/R + vehicle group at corresponding time point. c Representative Western blot images of Keap1 expression in the nucleus and cytoplasm after 8 h of OGD/R. Lamin A or β-actin was used as an internal control in the nucleus or cytoplasm, respectively. d Quantitative analysis of Keap1 levels in c (normalized to control for each part). * P< 0.05 vs. control group. All data were presented as mean ± SD of triplicate independent experiments. One-way ANOVA analysis followed by Newman–Keuls test
To determine whether the increased nuclear accumulation caused by Rg1 is regulated by Keap1, its expression was determined at 24 h of OGD/R. As shown in Fig. 2c, d, the expression of Keap1 in both the nucleus and the cytoplasm was significantly decreased after OGD/R, but Rg1 treatment showed no effect on Keap1 expression compared with the vehicle group. This suggests that Keap1 does not play a role in the Rg1-mediated enhancement of the nuclear accumulation of Nrf2.
Rg1 increases Nrf2-targeted gene expression
To determine whether ARE activity is increased by Rg1, ARE-dependent luciferase activity was evaluated. As shown in Fig. 3a, Rg1 treatment increased ARE-luciferase activity in a concentration-dependent manner (P < 0.01, P < 0.001 vs vehicle group). We next tested the ability of Rg1 to promote the Nrf2 pathway under OGD/R. As shown in Fig. 3b, quantitative PCR analysis showed that mRNA levels of NQO-1 (P < 0.01, P < 0.001 vs model group), HO-1 (P < 0.05, P < 0.01 vs model group), GCLC (P < 0.01, P < 0.001 vs model group), and GCLM (P < 0.05, P < 0.01 vs model group) are significantly increased after Rg1 treatment, which were alleviated in transcription caused by OGD/R. Furthermore, the protein expression of the Nrf2 pathway was also investigated. Fig. 3c, d show that Rg1 increased the levels of expression of NQO-1, HO-1, GCLC, GCLM significantly at the concentrations of 0.10 μM and 1.00 μM. Collectively, the results suggest that Rg1 stimulates ARE activity in OGD/R and subsequently upregulates the expression of target mRNAs and proteins.
Rg1 contributes to ARE-targeted gene expressions. a Rg1-induced ARE-dependent luciferase activity. ARE-luciferase reporter was transfected into PC12 cells by Lipofectamine 2000 for 48 h, then, they were treated with vehicle or Rg1 for the next 8 h at indicated concentration. Luciferase activity was determined by an analyzer fluorescent assay (* P< 0.05, **P < 0.01, ***P < 0.001 vs. vehicle group). b Rg1 promoted ARE-regulated gene transcription after OGD/R. The mRNA levels were tested by quantitative RT-PCR, including NQO-1, HO-1, GCLC, and GCLM (#P < 0.05 vs. control group, *P < 0.05, **P < 0.01, ***P < 0.001 vs. model group). c Rg1 promoted ARE-regulated protein expression after OGD/R. The levels of NQO-1, HO-1, GCLC, and GCLM were tested by Western blot. d Quantitative analysis of HO-1, GCLC, GCLM, and NQO-1 protein levels (#P < 0.05 vs. control group,*P < 0.05, **P < 0.01, ***P < 0.001 vs. model group). All data were presented as mean ± SD of triplicate independent experiments. One-way ANOVA analysis followed by Newman–Keuls test
Rg1 improves OGD/R-induced neurotoxicity in a Nrf2-dependent manner
To investigate whether the neuroprotective effect of Rg1 acts via Nrf2 activation, Nrf2-siRNA was employed to block the expression of Nrf2. As shown in Fig. 4a, b, Nrf2-siRNA abolished the increase in nuclear accumulation of Nrf2 induced by Rg1 after OGD. Correspondingly, the ARE-luciferase activity and Nrf2 pathway were no longer enhanced by Rg1 as shown in Fig. 4c–e. We also found that Nrf2-siRNA silenced the neuroprotective effect of Rg1 after OGD/R, as exhibited by decreased cell viability and increased ROS levels (Fig. 4f, h). The above results demonstrated that the anti-OGD/R effect of Rg1 is dependent on the activation of Nrf2.
Rg1 improves OGD/R-induced neurotoxicity in an Nrf2-dependent manner in vitro. a Representative Western blot images for PC12 cells transfected with control-siRNA or Nrf2-siRNA, with or without Rg1 (1.00 μM) treatment. Lamin A was used as an internal control.b Quantitative analysis of Nrf2 level in a. (***P < 0.001 vs. vehicle; ns: no significant difference). c Nrf2-siRNA blocked the increased ARE-luciferase activity induced by Rg1 after OGD/R in PC12 cells. Luciferase activity was determined by an analyzer fluorescent assay. (***P < 0.001 vs. vehicle; ns: no significant difference). d Representative Western blot images of ARE-targeted proteins expression, including NQO-1, HO-1, GCLC, and GCLM, induced by Rg1 when Nrf2 expression was normal or blocked. e Quantitative analysis of the levels of expression of NQO-1, HO-1, GCLC, and GCLM in d. (**P < 0.01 vs. vehicle; ns: no significant difference). f Cell viability was determined by MTT assay. (###P < 0.001 vs. control,** P< 0.01 vs. model; ns: no significant difference). g The released LDH from PC12 cells was determined by colorimetry. (###P < 0.001 vs. control, *P < 0.05 vs. model; ns: no significant difference). h The level of intracellular ROS was also revealed by DCFH-DA staining. (##P < 0.01 vs. control, * P< 0.05 vs. model; ns: no significant difference). All data were expressed as mean ± SD from three independent experiments. One-way ANOVA analysis followed by Newman–Keuls test
MiR-144 mediated the increased Nrf2 transcriptional activity by Rg1 under OGD
As previously mentioned, Rg1 had no effect on the expression of Keap1, though Nrf2 accumulation was enhanced significantly. It has been well documented that posttranscriptional regulation via microRNAs (miRNAs), such as miR-153, miR-144, and others, plays an important role in Nrf2/ARE activity in the central nervous system [33, 40]. In this study, qPCR evaluation showed that OGD/R raised miR-144 expression, while Rg1 treatment reversed it significantly (P < 0.01 vs model group, Fig. 5a). However, the expression of miR-144 is unaffected by the inhibition of Nrf2 expression by its siRNA, indicating that miR-144 is not regulated by Nrf2. It has been documented that miR-144 directly regulates Nrf2 and modulates the oxidative stress response in K562 cells [41]. To determine whether miR-144 could regulate Nrf2 activation directly in neurons, scramble control or miR-144 were transfected into PC12 cells and the nuclear and cytoplasmic accumulation of Nrf2 and ARE-luciferase activity were tested. As shown in Fig. 5b–e, miR-144 reduced both nuclear and cytoplasmic accumulation of Nrf2 compared to scramble control transfection (P < 0.001 vs scramble control-treated cells). Figure 5f showed that miR-144 transfection inhibited ARE-luciferase significantly (P < 0.001 vs scramble control-treated cells), which is consistent with findings in K562 cells [38].
MiR-144 mediates the increased Nrf2 transcriptional activity by Rg1 under OGD/R. a PC12 cells were treated with vehicle or Rg1 after 24 h followed with OGD, miRNA-144 level was determined by real-time PCR, and U6 was used as an internal control. Data were expressed as relative intensity of miR-144 normalized to U6 over control group. (** P< 0.01 vs. model). b, d Representative Western blot images of nuclear and cytoplasmic Nrf2 expression, respectively, after transfected with miR-144 or scramble control. β-actin or Lamin A was used as an internal control in the cytoplasm or nucleus, respectively. c, e are the quantitative analyses of b, d, respectively (*** P< 0.001 vs. scramble control).f ARE-luciferase activity after transfection with miR-144 or scramble control. ARE-luciferase was normalized to renilla activity, and the result was normalized to scramble control (*** P< 0.001 vs. scramble control). g The binding site between miR-144 and Nrf2 as predicted by TargetScan at 349-356 nt of 3’-UTR in Nrf2. Nrf2-3’-UTR-MUT was constructed with three mutations in the miR-144-binding site at 352, 353, and 354 as shown in the bottom line of (g, h) PC12 cells were co-transfected with reporter gene constructs and pRL-renilla constructs with scramble control or miR-144. In addition, OGD was performed in the cells transfected with scramble followed by vehicle or Rg1 (1.00 μM) treatment for 24 h. Luciferase activity was normalized to renilla activity. Results were represented as normalized to control for Nrf2-3’-UTR or Nrf2-3’-UTR-MUT, respectively. (** P< 0.01). i Nrf2 3’-UTR construct was co-transfected with control anti-miR or anti-miR-144 into PC12 cells, followed by OGD/R, and then the luciferase activity was measured. (***P < 0.001 vs. control, ns: no significant difference). j ARE-luciferase activity results after co-transfection with control-anti-miR or anti-miR-144 followed by OGD/R. (**P < 0.01 vs. control, ns: significant difference). All data were expressed as mean ± SD from three independent experiments. One-way ANOVA analysis followed by Newman–Keuls test was used for a, h, i, j. Student’s t test was used for testing between two groups in c, e, f
According to TargetScan (www.targetscan.org), the 3’-untranslated region (3’-UTR) of Nrf2 is targeted by miR-144. To test whether the OGD/R-induced increased miR-144 directly targeted Nrf2 by binding to the 3’-UTR sequence, two reporters (Nrf2-3’UTR and miR-144-binding site mutated, Nrf2-3’-UTR-MUT) were constructed as shown in Fig. 5g. Both constructs were transfected into PC12 cells, followed by exposure to OGD/R with or without Rg1. MiR-144 was introduced as a positive control. As shown in Fig. 5h, OGD/R decreased luciferase activity in the Nrf2-3’-UTR group, (P < 0.01 vs control group), which is similar to the result in miR-144 exposure. Rg1 treatment was able to reverse this significantly (P < 0.01 vs OGD/R group). However, OGD/R showed no effect on the luciferase in Nrf2-3’-UTR-MUT group. As a positive control, miR-144 loses its inhibition on luciferase activity. These findings indicate that miR-144 blocks Nrf2 activity by targeting at the 3’-UTR of Nrf2. To determine whether OGD/R-induced decreased Nrf2 activity is due to miR-144, anti-miR-144, or control-anti-miR were transfected into PC12 cells along with Nrf2-3’-UTR, respectively. The results in Fig. 5i show that there is significantly decreased luciferase activity in control-anti-miR-treated cells after OGD/R. When miR-144 was absent owing to the anti-miR-144 transfection, OGD/R was unable to affect the luciferase activity. These results indicate that the repression of Nrf2 activity by OGD/R was due to the increase of miR-144 specifically. To further investigate the role of miR-144 on ARE luciferase, control-anti-miR or anti-miR-144 were also introduced into PC12 cells, respectively. We found that anti-miR-144 eliminated the decrease of ARE-luciferase induced by OGD/R (Fig. 5j). These findings further indicate that miR-144 suppressed Nrf2 transcriptional activity in PC12 cells after OGD/R.
Blockage of miR-144 silenced the anti-I/R effect of Rg1 in vivo
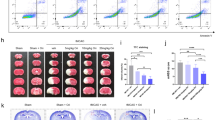
To further investigate a potential mechanistic role for miR-144 in the anti-I/R effect of Rg1 in vivo, we injected miR-144-shRNA in the predicted ischemic penumbra zone [31, 32] followed by tMCAO in SD rats to block the increased expression of miR-144 post-I/R. The result showed that Rg1 treatment at the dose of 2 mg/kg improved I/R outcomes in NC animals significantly (Fig. 6a–d), as manifested by decreased infarct volume (P < 0.05, Fig. 6a, b) and improved Longa’s score (P < 0.05, Fig. 6c) compared with the vehicle group. However, both of these improvements were erased by miR-144-shRNA administration, suggesting that miR-144 plays pivotal role in the anti-I/R effect of Rg1 in vivo.
Blockage of miR-144 abolished the anti-I/R effect of Rg1 in vivo. a Representative TTC staining images of tMCAO rats treated with vehicle or Rg1 (20 mg/kg) infected by miR-144 or control virus (NC) in ischemic penumbra. b Quantitative analysis of cerebral infarct volume in a. (*P < 0.05 vs. vehicle, ns: no significant difference, n = 8–10). c Quantitative analysis of neurological score by the Longa test. (* P< 0.05 vs. vehicle, ns: no significant difference, n = 8–10). d, e The level of intracellular ROS by DCFH-DA staining in cerebral cortex. e Representative images of ROS contents labeled in red, and d is the quantitative analysis of e (* P< 0.05 vs. model; ns: no significant difference). f, g The level of DNA oxidative product (8-OHDG) revealed by immunofluorescence staining in cerebral cortex. f Representative images of 8-OHDG staining shown in green. g Quantitative analysis of f (**P < 0.01 vs. model; ns: no significant difference). All data were expressed as mean ± SD from three independent experiments d–g. One-way ANOVA analysis followed by Newman–Keuls test
We then investigated whether the improved outcomes are associated with the changes of oxidative stress status. Based on the ROS evaluation using DCFH-DA, we found that Rg1 administration decreased intracellular levels of ROS significantly (P < 0.05 vs vehicle-treated animals in NC group) and was erased by miR-144-shRNA administration (Fig. 6d, e). Furthermore, the fluorescence intensity of 8-OHDG staining was used to measure the amount of DNA oxidation products. As shown in Fig. 6f, g, the fluorescence intensity of 8-OHDG was significantly decreased by Rg1 treatment in NC group (P < 0.01 vs vehicle-treated animals), However, administration of miR-144-shRNA reduced the level of 8-OHDG remarkably when compared with that in NC-treated animals. All of these results suggested that inhibition of miR-144 blocks the anti-I/R effect of Rg1 in vivo.
Inhibition of miR-144 mimics the enhancement of Nrf2/ARE signaling pathway induced by Rg1 after I/R
The anti-I/R effect of Rg1 was significantly reduced in the absence of miR-144, which could interact with the 3’-UTR of Nrf2 directly in vitro. To determine whether miR-144 produces similar biological activities in vivo, the level of Nrf2 and its target genes were determined by qPCR and Western blotting 24 h after tMCAO. As shown in Fig. 7a, b, there is a significant increase in nuclear accumulation of Nrf2 after Rg1 treatment compared with the vehicle in NC animals. However, the increase induced by Rg1 was similar with the administration of miR-144-shRNA, suggesting that the inhibition of miR-144 in vivo has a similar effect on the nuclear accumulation of Nrf2 in vitro. With the increase of nuclear Nrf2, its target genes were also upregulated. As shown in Fig. 7c–g, the Western blot result showed that Rg1 treatment increased the expression of NQO-1 (P < 0.05), HO-1 (P < 0.05), GCLC (P < 0.01), and GCLM (P < 0.01) significantly in NC animals. Application of miR-144-shRNA promotes the expression of these proteins compared with that in NC animals but abolishes the improvement induced by Rg1 treatment. Quantitative PCR determination revealed similar changes with proteins correspondingly in Fig. 7h–k, showing no significant difference in expression after shRNA transfection with or without Rg1 treatment. All of these results suggested that miR-144 is required for the enhancement of the Nrf2/ARE pathway induced by Rg1 in I/R injury.
Inhibition of miR-144 blocked the enhancement of Nrf2/ARE pathway induced by Rg1 after I/R. a Representative Western blot images of nuclear Nrf2 isolated from rat predicted penumbra cortex treated with vehicle or Rg1 (20 mg/kg) and transfected with control virus (NC) or miR-144-shRNA. Lamin A was used as an internal control. b Quantitative analysis of Nrf2 levels in a. (*P < 0.05 vs. vehicle; ns: no significant difference). c Representative images of Nrf2-targeted protein expressions isolated from predicted penumbra cortex treated with vehicle or Rg1 (20 mg/kg) and transfected with control virus (NC) or miR-144-shRNA. β-actin was used as an internal control. d–g Quantitative analysis of Nrf2-targeted gene expressions in c, including NQO-1 d, HO-1 e, GCLC f, and GCLM g, normalized to vehicle-treated animals in each NC infection. (* P< 0.05, ** P< 0.01 vs. vehicle; ns: no significant difference). h–k Quantitative analysis of Nrf2-targeted mRNA levels determined by qPCR in the predicted penumbra cortex, including NQO-1 h, HO-1 i, GCLC j, and GCLM k, normalized to vehicle-treated animals in each NC infection. (* P< 0.05, ** P< 0.01 vs. vehicle; ns: no significant difference). All data were expressed as mean ± SD from three independent experiments. One-way ANOVA analysis followed by Newman–Keuls test
Discussion
Stroke is one of the leading causes of human death and disability [14] and lacks effective treatment. Rg1 is well documented to have a beneficial effect on the I/R-mediated neurotoxicity often experienced during strokes [6]. In this study, we found that Rg1 protects against I/R-induced neurotoxicity by improving nrf2 activity both in vivo and in vitro. We also found that the neuroprotective effect of Rg1 is dependent on the miR-144-Nrf2/ARE pathway, which provides a new clue for the treatment of ischemic stroke.
The neuroprotective actions of Rg1 have been documented to have beneficial effects in experimental stroke therapy, including improved neurological function, reduced infarct volume, improved BBB integrity, and other supporting evidence [6]. It has been proven that Rg1 protects the brain against oxidative stress in many pathological conditions, such as Parkinson’s disease [42, 43], Alzheimer disease [44], d-galactose-induced brain aging [45], and others. Our previous study showed that Rg1’s hepatic protection effect was mainly due to its antioxidant activity [35, 46]. However, whether the efficacy of Rg1 against oxidative stress is involved in its antistroke capability has not been reported until now. To test this hypothesis, we treated PC12 cells with Rg1 or vehicle followed by OGD. We found that Rg1 significantly protects neurons against OGD/R from 0.01 μM to 1.00 μM in a concentration-dependent manner, as revealed by improved neural viability and reduced LDH release, displayed in Fig. 1b, c, we found that 1.00 μM exhibited the best therapeutic effect. When the concentration of Rg1 increased to 10.0 μM, the protective effect of Rg1 dropped; hence, the high concentration of Rg1 was not extensively investigated in this study. The lowest concentration of Rg1 in this study was 0.01 μM; Fig. 1c, d show that it is ineffective for reducing the level of LDH and production of ROS, the most direct stimulator for Nrf2, suggesting that the neuroprotective effect of Rg1 at the concentration of 0.01 μM may be unrelated to Nrf2. Therefore, higher or lower concentrations of Rg1 were not further investigated in this study.
It has been widely confirmed that I/R triggers a rapid and excessive ROS production, which promotes oxidative injury by lipid peroxidation, DNA peroxidation, etc. [11]. To test whether the neuroprotective effect of Rg1 is related to ROS, DCFH-DA staining was used to determine the level of intracellular ROS. We found that Rg1 treatment significantly reversed the post-I/R increase of ROS; likewise, the protective effect was exhibited in a concentration-dependent manner at a concentration between 0.10 μM and 1.00 μM (Fig. 1d). This finding is consistent with its efficacious concentration capable of reducing LDH release. These results suggest that the neuroprotective effect of Rg1 may be related to its ability to suppress excessive ROS release triggered by oxidative stress.
Oxidative stress occurs when intracellular ROS production overcomes intracellular antioxidative stress defenses [40], including enzymatic and non-enzymatic. Nrf2 is one of the master regulators of the expression of proteins involved in endogenous antioxidation defense [47], which is activated by excessive ROS release following IR injury. A number of studies have indicated that the expression of Nrf2 is largely increased in the acute phase of strokes [47]. Moreover, the change in Nrf2 level is mainly in neurons [21]. In our study, we found that Nrf2 nuclear accumulation induced by OGD/R in PC12 cells, begins at 2 h of OGD, peaks at 4 h, and decreases below baseline at ~ 8 h (Fig. 2a, b). Rg1 treatment significantly enhanced Nrf2 nuclear accumulation, as evidenced by a tendency of slower decline, going from 8 h and lasting for 24 h, which provides a solid basis for the promotion of antioxidation gene expression.
It is well known that the distribution and degradation of Nrf2 are regulated by its endogenous inhibitor Keap1 [46]. Keap1 is a homo-dimer with two canonical domains: one binds to Nrf2 and the other binds to E3 ubiquitin ligase complex, which keeps Nrf2 in the cytoplasm and promotes its degradation [19]. To determine the reason for increased nuclear accumulation of Nrf2 after I/R, the changes of Keap1 were investigated. As shown in Fig. 2c, d, we found that OGD/R decreased the Keap1 content in both the cytoplasm and the nucleus. Rg1 administration exhibited no effect on Keap1 expression, suggesting that the enhanced nuclear accumulation of Nrf2 induced by Rg1 may be a result of increased Nrf2 expression, rather than Keap1 regulation.
Nuclear Nrf2 forms heterodimers with bZIP proteins, recognizing the appropriate ARE sequences and then initiating the transcription of antioxidation genes harboring ARE, such as NQO-1 and HO-1, in their promoter region [23]. As the result of enhanced nuclear accumulation, we found that ARE-luciferase activity is enhanced by Rg1 directly (Fig. 3a), and then the activities of its downstream antioxidation genes and proteins are increased accordingly (Fig. 3b–d). All of these results suggested that the Nrf2/ARE pathway is activated by Rg1 after OGD/R. Nrf2-siRNA was used to investigate the role of Nrf2 in the anti-OGD/R effect of Rg1. Our results showed that Nrf2-siRNA significantly reduced Rg1’s neuroprotective effect in terms of ARE-luciferase activity, expression of downstream proteins, cell viability, LDH release, and ROS levels (Fig. 4c–h). These results indicate that the anti-OGD/R effect of Rg1 is in a Nrf2-dependent manner.
As previously mentioned, there is a significantly increased Nrf2 protein level compared with its mRNA contents, indicating that the increased nuclear portion of Nrf2 is owing to posttranscriptional regulation, such as by miRNAs. A number of miRNAs, such as miR-153 in the PD model [31, 40], miR-144 in the AD model [33] have been reported for their involvement in the regulation of Nrf2. In this study, we found that the level of miR-144 increased significantly after OGD/R, which exacerbated oxidative stress by inhibiting Nrf2 expression by targeting its 3’-UTR (Fig. 5a). MiR-144 was able to inhibit both the nuclear and cytoplasmic Nrf2 expression (Fig. 5b–e) and ARE-luciferase activity (Fig. 5f). Rg1 treatment reversed this effect significantly by releasing Nrf2 mRNA from miR-144 and promoting Nrf2 expression, shown in Fig. 5a. Moreover, we found that the expression of miR-144 was independent of Nrf2 activation in PC12 cells induced by OGD/R, as Nrf2-siRNA had no effect on the level of miR-144, shown in Fig. 5a. However, application of anti-miR-144 increased ARE-luciferase activity significantly (Fig. 5j), suggesting that miR-144 is an upstream regulator of Nrf2 expression. Furthermore, we found that miR-144 regulates Nrf2 by interacting with its 3’-UTR directly in PC12 cells, as shown in Fig. 5h. MiR-144 inhibited luciferase activity in the PC12 cells transfected with Nrf2-3’-UTR after OGD/R, but fails to do so in Nrf2-3’-UTR-MUT transfected cells. Rg1 treatment decreased the level of miR-144 significantly after OGD/R, which is also independent of Nrf2 level. Rg1’s ability to improve Nrf2 transcriptional activity is lost when Nrf2 3’-UTR was mutated in miR-144 seed sites. This suggests that miR-144 is the mediator of Rg1-induced activation of Nrf2/ARE pathway in vitro because Rg1 was no longer capable of promoting Nrf2 activity when the interaction between miR-144 and Nrf2 was inhibited.
Subsequently, we tested the role of miR-144 in the anti-I/R effect of Rg1 in vivo. Rg1 treatment improved the neural outcomes of tMCAO significantly at the dose of 20 mg/kg in NC-treated animals, including decreased infarct volume (Fig. 6a) and improved neurological scores (Fig. 6b), which are consistent with a previous study [7]. However, the application of miR-144-shRNA abolished these improvements significantly. The improved oxidative stress status mediated by Rg1, including lower intracellular ROS content (Fig. 6c) and DNA oxidative products (Fig. 6d–g), was also counteracted in miR-144-shRNA treated animals. Furthermore, we found that Rg1’s positive effect on the Nrf2/ARE pathway in I/R-induced neuron injury, including higher nuclear accumulation and target gene transcription and expression (Fig. 7), was also diminished by miR-144-shRNA administration. All of these results suggest that miR-144 plays a crucial role in mediating the Nrf2/ARE pathway in the anti-I/R effect of Rg1 in vivo.
Conclusions
In summary, we identified that the anti-I/R effect of Rg1 involves the Nrf2/ARE pathway and the underlying mechanism was clarified with posttranscriptional regulation of Nrf2 by miR-144. Furthermore (Fig. 8), we provide solid evidence for the direct regulatory effect of miR-144 on the Nrf2/ARE pathway in neurons, and confirmed that the enhanced Nrf2/ARE pathway mediated by Rg1 in I/R is dependent on miR-144. All of these results provide strong evidence in support of Rg1 as a promising drug target for the treatment of ischemic stroke.
Schematic diagram of the anti-I/R effect of Rg1 through miR-144/Nrf2/ARE pathway. I/R induces overproduction of ROS, which triggers oxidative stress and exhaust intracellular antioxidants. Meanwhile, it promotes antioxidants production by promoting the Nrf2/ARE pathway. I/R also promotes the activity of miR-144, which directly binds to Nrf2-3’-UTR in neurons and inhibits the expression of Nrf2. Rg1 inhibits the activity of miR-144 after I/R and promotes the Nrf2/ARE pathway, which enhances the expression of ARE-regulated antioxidative genes, reduces oxidative damage and exerts anti-I/R effect
Change history
29 April 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41401-024-01272-1
References
Wong AS, Che CM, Leung KW. Recent advances in ginseng as cancer therapeutics: a functional and mechanistic overview. Nat Prod Rep. 2015;32:256–72.
Shin EJ, Shin SW, Nguyen TT, Park DH, Wie MB, Jang CG, et al. Ginsenoside Re rescues methamphetamine-induced oxidative damage, mitochondrial dysfunction, microglial activation, and dopaminergic degeneration by inhibiting the protein kinase Cdelta gene. Mol Neurobiol. 2014;49:1400–21.
Wang J, Li D, Hou J, Lei H. Protective effects of geniposide and ginsenoside Rg1 combination treatment on rats following cerebral ischemia are mediated via microglial microRNA1555p inhibition. Mol Med Rep. 2018;17:3186–93.
Li Y, Suo L, Liu Y, Li H, Xue W. Protective effects of ginsenoside Rg1 against oxygen-glucose-deprivation-induced apoptosis in neural stem cells. J Neurol Sci. 2017;373:107–12.
Zhou Y, Li HQ, Lu L, Fu DL, Liu AJ, Li JH, et al. Ginsenoside Rg1 provides neuroprotection against blood brain barrier disruption and neurological injury in a rat model of cerebral ischemia/reperfusion through downregulation of aquaporin 4 expression. Phytomedicine. 2014;21:998–1003.
Xie CL, Wang WW, Xue XD, Zhang SF, Gan J, Liu ZG. A systematic review and meta-analysis of Ginsenoside-Rg1 (G-Rg1) in experimental ischemic stroke. Sci Rep. 2015;5:7790.
Lin M, Sun W, Gong W, Ding Y, Zhuang Y, Hou Q. Ginsenoside Rg1 protects against transient focal cerebral ischemic injury and suppresses its systemic metabolic changes in cerabral injury rats. Acta Pharm Sin B. 2015;5:277–84.
Shimizu Y, Nicholson CK, Lambert JP, Barr LA, Kuek N, Herszenhaut D, et al. Sodium sulfide attenuates ischemic-induced heart failure by enhancing proteasomal function in an Nrf2-dependent manner. Circ Heart Fail. 2016;9:e002368.
Wakayama K, Shimamura M, Suzuki JI, Watanabe R, Koriyama H, Akazawa H, et al. Angiotensin II peptide vaccine protects ischemic brain through reducing oxidative stress. Stroke. 2017;48:1362–8.
Li P, Shen M, Gao F, Wu J, Zhang J, Teng F, et al. An antagomir to MicroRNA-106b-5p ameliorates cerebral ischemia and reperfusion Injury in rats via inhibiting apoptosis and oxidative stress. Mol Neurobiol. 2017;54:2901–21.
Li P, Stetler RA, Leak RK, Shi Y, Li Y, Yu W, et al. Oxidative stress and DNA damage after cerebral ischemia: Potential therapeutic targets to repair the genome and improve stroke recovery. Neuropharmacology. 2017;134:208–17.
Fraser PA. The role of free radical generation in increasing cerebrovascular permeability. Free Radic Biol Med. 2011;51:967–77.
Radermacher KA, Wingler K, Langhauser F, Altenhofer S, Kleikers P, Hermans JJ, et al. Neuroprotection after stroke by targeting NOX4 as a source of oxidative stress. Antioxid Redox Signal. 2013;18:1418–27.
Zheng Y, Wang Y, Zhang X, Tan Y, Peng S, Chen L, et al. C19, a C-terminal peptide of CKLF1, decreases inflammation and proliferation of dermal capillaries in psoriasis. Sci Rep. 2017;7:13890.
Carbone F, Teixeira PC, Braunersreuther V, Mach F, Vuilleumier N, Montecucco F. Pathophysiology and treatments of oxidative injury in Ischemic stroke: focus on the phagocytic NADPH oxidase 2. Antioxid Redox Signal. 2015;23:460–89.
Takayasu Y, Nakaki J, Kawasaki T, Koda K, Ago Y, Baba A, et al. Edaravone, a radical scavenger, inhibits mitochondrial permeability transition pore in rat brain. J Pharmacol Sci. 2007;103:434–7.
Tang M, Feng W, Zhang Y, Zhong J, Zhang J. Salvianolic acid B improves motor function after cerebral ischemia in rats. Behav Pharmacol. 2006;17:493–8.
Wu G, Zhu L, Yuan X, Chen H, Xiong R, Zhang S, et al. Britanin ameliorates cerebral ischemia-reperfusion injury by inducing the Nrf2 protective pathway. Antioxid Redox Signal. 2017;27:754–68.
Xu X, Zhang L, Ye X, Hao Q, Zhang T, Cui G, et al. Nrf2/ARE pathway inhibits ROS-induced NLRP3 inflammasome activation in BV2 cells after cerebral ischemia reperfusion. Inflamm Res. 2018;67:57–65.
Wang Y, Huang Y, Xu Y, Ruan W, Wang H, Zhang Y, et al. A dual AMPK/Nrf2 activator reduces brain inflammation after stroke by enhancing microglia M2 polarization. Antioxid Redox Signal. 2018;28:141–63.
Tanaka N, Ikeda Y, Ohta Y, Deguchi K, Tian F, Shang J, et al. Expression of Keap1-Nrf2 system and antioxidative proteins in mouse brain after transient middle cerebral artery occlusion. Brain Res. 2011;1370:246–53.
Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011;108:1427–32.
Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. J Neurosci. 2005;25:10321–35.
Alfieri A, Srivastava S, Siow RC, Cash D, Modo M, Duchen MR, et al. Sulforaphane preconditioning of the Nrf2/HO-1 defense pathway protects the cerebral vasculature against blood-brain barrier disruption and neurological deficits in stroke. Free Radic Biol Med. 2013;65:1012–22.
Sandberg M, Patil J, D’Angelo B, Weber SG, Mallard C. NRF2-regulation in brain health and disease: implication of cerebral inflammation. Neuropharmacology. 2014;79:298–306.
Ding Y, Chen M, Wang M, Li Y, Wen A. Posttreatment with 11-keto-beta-boswellic acid ameliorates cerebral ischemia-reperfusion injury: Nrf2/HO-1 pathway as a potential mechanism. Mol Neurobiol. 2015;52:1430–9.
Guo Y, Yu S, Zhang C, Kong AN. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med. 2015;88:337–49.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31.
Yu M, Liu Y, Zhang B, Shi Y, Cui L, Zhao X. Inhibiting microRNA-144 abates oxidative stress and reduces apoptosis in hearts of streptozotocin-induced diabetic mice. Cardiovasc Pathol. 2015;24:375–81.
Liu L, Sun T, Liu Z, Chen X, Zhao L, Qu G, et al. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS One. 2014;9:e103948.
Ji Q, Gao J, Zheng Y, Liu X, Zhou Q, Shi C, et al. Inhibition of microRNA-153 protects neurons against ischemia/reperfusion injury in an oxygen-glucose deprivation and reoxygenation cellular model by regulating Nrf2/HO-1 signaling. J Biochem Mol Toxicol 2017; 31. https://doi.org/10.1002/jbt.21905.
Li J, Rohailla S, Gelber N, Rutka J, Sabah N, Gladstone RA, et al. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res Cardiol. 2014;109:423.
Zhou C, Zhao L, Zheng J, Wang K, Deng H, Liu P, et al. MicroRNA-144 modulates oxidative stress tolerance in SH-SY5Y cells by regulating nuclear factor erythroid 2-related factor 2-glutathione axis. Neurosci Lett. 2017;655:21–7.
Chu SF, Zhang Z, Zhang W, Zhang MJ, Gao Y, Han N, et al. Upregulating the expression of survivin-HBXIP complex contributes to the protective role of IMM-H004 in transient global cerebral ischemia/reperfusion. Mol Neurobiol. 2017;54:524–40.
Gao Y, Chu S, Shao Q, Zhang M, Xia C, Wang Y, et al. Antioxidant activities of ginsenoside Rg1 against cisplatin-induced hepatic injury through Nrf2 signaling pathway in mice. Free Radic Res. 2017;51:1–13.
Ashwal S, Tone B, Tian HR, Cole DJ, Pearce WJ. Core and penumbral nitric oxide synthase activity during cerebral ischemia and reperfusion. Stroke. 1998;29:1037–46.
Ashwal S, Tone B, Tian HR, Cole DJ, Liwnicz BH, Pearce WJ. Core and penumbral nitric oxide synthase activity during cerebral ischemia and reperfusion in the rat pup. Pediatr Res. 1999;46:390–400.
Niu F, Song XY, Hu JF, Zuo W, Kong LL, Wang XF, et al. IMM-H004, a new coumarin derivative, improved focal cerebral ischemia via blood-brain barrier protection in rats. J Stroke Cerebrovasc Dis. 2017;26:2065–73.
Kong LL, Wang ZY, Han N, Zhuang XM, Wang ZZ, Li H, et al. Neutralization of chemokine-like factor 1, a novel C-C chemokine, protects against focal cerebral ischemia by inhibiting neutrophil infiltration via MAPK pathways in rats. J Neuroinflammation. 2014;11:112.
Zhang XS, Ha S, Wang XL, Shi YL, Duan SS, Li ZA. Tanshinone IIA protects dopaminergic neurons against 6-hydroxydopamine-induced neurotoxicity through miR-153/NF-E2-related factor 2/antioxidant response element signaling pathway. Neuroscience. 2015;303:489–502.
Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–48.
Heng Y, Zhang QS, Mu Z, Hu JF, Yuan YH, Chen NH. Ginsenoside Rg1 attenuates motor impairment and neuroinflammation in the MPTP-probenecid-induced parkinsonism mouse model by targeting alpha-synuclein abnormalities in the substantia nigra. Toxicol Lett. 2016;243:7–21.
Chen XC, Zhou YC, Chen Y, Zhu YG, Fang F, Chen LM. Ginsenoside Rg1 reduces MPTP-induced substantia nigra neuron loss by suppressing oxidative stress. Acta Pharmacol Sin. 2005;26:56–62.
Yu HT, Zhen J, Pang B, Gu JN, Wu SS. Ginsenoside Rg1 ameliorates oxidative stress and myocardial apoptosis in streptozotocin-induced diabetic rats. J Zhejiang Univ Sci B. 2015;16:344–54.
Li J, Cai D, Yao X, Zhang Y, Chen L, Jing P, et al. Protective effect of ginsenoside Rg1 on hematopoietic stem/progenitor cells through attenuating oxidative stress and the Wnt/beta-catenin signaling pathway in a mouse model of d-galactose-induced aging. Int J Mol Sci. 2016;17:pii:E849.
Li JP, Gao Y, Chu SF, Zhang Z, Xia CY, Mou Z, et al. Nrf2 pathway activation contributes to anti-fibrosis effects of ginsenoside Rg1 in a rat model of alcohol- and CCI4-induced hepatic fibrosis. Acta Pharmacol Sin. 2014;35:1031–44.
Zhang R, Xu M, Wang Y, Xie F, Zhang G, Qin X. Nrf2-a promising therapeutic target for defensing against oxidative stress in stroke. Mol Neurobiol. 2017;54:6006–17.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81603315, 81730096, 81603316, 81503275, 81730093, 81873026, U1402221), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-004), the State Key Laboratory Fund Open Project (GTZK201610), the China Postdoctoral Science Foundation (2013M540066), the PUMC Graduate Education and Teaching Reform Project (10023201600801), the Beijing Key Laboratory of New Drug Mechanisms and Pharmacological Evaluation Study (BZ0150), the opening Program of Shanxi Key Laboratory of Chinese Medicine Encephalopathy (CME-OP-2017001), and the Hunan Chinese Herbal Medicine and standardization functions Engineering Research Center (BG201701).
Author information
Authors and Affiliations
Contributions
N.-h.C. designed research; S.-f.C. and Z.Z. performed the cell experiments and Nrf2-related determination; Xin Zhou, Chen Chen, and Dan-dan Liu prepared the animal models; W.-B.H. guided the animal experiments; P.L., Z.-z.W. and Q.-d.A. determined ROS and 8-OHDG in brain slide; S.-f.C., H.-f.G., H.-S.S. and Z.-P.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Chu, Sf., Zhang, Z., Zhou, X. et al. Ginsenoside Rg1 protects against ischemic/reperfusion-induced neuronal injury through miR-144/Nrf2/ARE pathway. Acta Pharmacol Sin 40, 13–25 (2019). https://doi.org/10.1038/s41401-018-0154-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-018-0154-z