Abstract
Circular RNAs (circRNAs), one kind of noncoding RNAs, can interact with miRNA and transcription factors to regulate gene expression. However, little is known on which circRNA is crucial for the pathogenesis of hepatocellular carcinoma (HCC). CircRNA expression profile was analyzed by a microarray. Regulatory gene targets were predicted by bioinformatics analysis and validated by luciferase assay. Their expression was determined by qRT-PCR and Western blotting. DNA methylation was determined by methylation-specific PCR. Gene knockdown and overexpression were mediated by lentivirus-mediated shRNA and transfection with plasmids for cDNA expression, respectively. MTT assay, wound-healing assay, transwell invasion assay, and flow cytometry were used to determine malignant behaviors of HCC cells. HCC xenograft mouse model was used to determine the in vivo effects of circRNA-5692. CircRNA-5692 expression was downregulated in HCC tissues, and circRNA-5692 overexpression attenuated the malignant behaviors of HCC cells. Bioinformatics predicted that circRNA-5692 interacted with miR-328-5p, which targeted the DAB2IP mRNA. Actually, miR-328-5p promoted the malignant behaviors of HCC cells, while DAB2IP had opposite effects. Moreover, circRNA-5692 overexpression inhibited the growth of xenograft HCC tumors in vivo by decreasing miR-328-5p expression to enhance DAB2IP expression. In conclusion, the circRNA-5692–miR-328-5p–DAB2IP regulatory pathway inhibits the progression of HCC. Our findings may provide potential new targets for the diagnosis and therapy of HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) accounts for 90% of primary liver carcinomas in the world1,2. HCC is one of the leading causes of malignancy in humans, and has high morbidity and mortality rates3. Despite of significant advance in therapeutic strategies for HCC, the 5-year survival rate of HCC patients remains low4. More importantly, little is known on the molecular pathogenesis and therapeutic targets of HCC. Therefore, understanding the pathogenic process of HCC and its regulatory mechanisms will be of great significance in management of HCC.
Circular RNAs (circRNAs), microRNAs (miRNAs), and other noncoding RNAs can regulate cell activities5. CircRNAs are covalently closed continuous loop type of single-stranded RNAs in mammalian cells ubiquitously and regulate gene expression6. Previous studies have shown that circRNAs can regulate malignant behaviors, including the proliferation, migration, invasion, and apoptosis of cancer cells7,8,9,10. For example, circRNA hsa_circ_0000096 regulates the proliferation and migration of gastric cancer cells by modulating the gene expression of cyclin D1, cyclin-dependent kinase-6 (CDK-6), matrix metalloproteinase-2 (MMP-2), MMP-9, and E-cadherin8. The circRNA PVT1 and hsa_circ_0067934 act as oncogenic factors to promote the proliferation of gastric cancer and esophageal carcinoma cells9,10. CircRNAs can act as competing endogenous RNAs (ceRNAs) to sponge their target miRNAs via direct binding to miRNAs, and modulate their activity, regulating the miRNA-targeted gene expression11,12. A recent study has indicated that circRNA CiRS-7 can promote the growth and metastasis of esophageal squamous cell carcinoma by sponging miR-7 and suppressing its activity13. The circRNA Vav3 can sponge gga-miR-375 to affect the YAP1 expression, by promoting the epithelial mesenchymal transition (EMT) process14. Furthermore, circRNAs can interact with transcription factors and other proteins to form circRNPs, and modulate their function15,16,17. A recent study suggests that circRNAs are important for the initiation, migration, and invasion of HCC18. However, which circRNA and how it regulates the malignant behaviors of HCC have not been clarified.
In this study, we screened differentially expressed circRNAs that were significantly downregulated in HCC tissues and identified that circRNA-5692 overexpression effectively attenuated the malignant behaviors of HCC. Furthermore, we explored its potential targeted miRNAs and found that circRNA-5692 acted as a ceRNA to sponge oncogenic miR-328-5p to enhance tumor suppressor DAB2IP (Ras (MIM 190020) GTPase-activating protein) expression, attenuating the malignant behaviors of HCC in vitro and in vivo.
Materials and methods
HCC specimens and microarray of circRNAs
The experimental protocols were approved by Ethics Committee of Xiangya Hospital of Central South University (201502012). We collected 92 pairs of surgical or biopsied HCC and adjacent non-tumor specimens in Xiangya Hospital of Central South University, Hunan Cancer Hospital, and Affiliated Hospital of Guilin Medical College from February 2017 to June 2018. All samples were collected with patient consent and signed informed consent. The HCC patients were diagnosed, based on the practice guidelines of the American Association for the Study of Liver Diseases (AASLD). Their liver specimens were evaluated by pathologists and their clinical stages were determined, according to the BCLC classification. HCC patients with the following conditions were excluded: (1) patients ≤ 18 or ≥ 70 years of age or without full civil capacity; (2) patients with a history of preoperative anticancer radiotherapy or chemotherapy, biological, immune, and traditional Chinese medicine; (3) patients with incomplete postoperative follow-up data; (4) patients with a history of another organ malignancy, or systemic immune disease. Their tissue RNA was extracted by using a RNeasy Mini Kit (Qiagen, Hilden, Germany), and the RNA concentrations were measured by a NanoDrop 1000 spectrophotometer (ThermoScientific, Waltham, MA). The contained circRNAs were enriched and digested with RNase A and reversely transcribed into cRNA by using fluorescent reagents and random primers. The circRNA profile was determined by Microarray by using Human circRNA Arrays (8 × 15 K, Arraystar, Rockville, MD, USA). A change of ≥ 2-fold in the circRNA level was defined as differential expression. The data and potential interaction with miRNAs were analyzed by using R software limma and Arraystar program (Arraystar).
Cell culture
Human HCC HCCLM3, Huh-7, HepG2, cervical cancer Chang Liver cells, non-tumor human embryotic kidney HEK293T, liver WR168, and LX-2 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in DMEM containing 20% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/ml of penicillin, and 100 µg/ml of streptomycin (complete medium, Gibco, Aukland, New Zealand) at 37 °C in an incubator of 5% CO2.
Transfection and transduction of cells
The plasmid pHBLV-CMV-Cicr-MCS-EF1-circRNA-5692, its derived lentivirus, miRNA-328-5p mimics, miRNA-328-shRNA, miR-1207-5p mimic, and their negative control miRNAs, their relevant miRNA-derived lentiviruses, plasmids for expression of DAB2IP, DAB2IP-specific shRNA, and their relevant lentiviruses were obtained from Shanghai Hanheng Biotech (Shanghai, China). HCC cells were transduced with individual types of lentivirus at a multiplicity of infection (MOI) of 10 in the presence of 5 µg/ml puromycin or transfected with the specific plasmid by using Lipofectamine 2000 (ThermoFisher).
Quantitative PCR and methylation-specific PCR
The different groups of cells were harvested and their total RNAs were extracted by using Trizol reagent (Invitrogen). The RNA samples (1–3 µg each) were reversely transcribed into cDNA by using the Superscrpt II kit (Invitrogen). The relative levels of target gene RNA transcripts were determined by qRT-PCR by using the SYBR Green mix (Kakara, Dalian, China) and specific primers in an ABI 7500 thermocycler (Applied Biosystems). The sequences of primers were forward 5′-GCCTGAATGATGACTGCTGA-3′ and reverse 5′-GGTAACAGAAGCGCCTGAAG-3′ for hcirc-0001727-220; Forward 5′-GCTCGACCTGAAGCTGAGTA-3′ and Reverse 5′- CTTGGAGTTCAGCAGGAAGC-3′ for hcirc-0005692-172; Forward 5′-CATTGCCCCATGTGAAGTC-3′ and Reverse 5′-GGTGCCCCTGGAGATTTTAG-3′ for hcirc-0028861-109; Forward 5′-GTATGGTGTGGCTTGTGTGG-3′ and Reverse 5′- GCTGCAATCCTCAGAGAAGG-3′ for hcirc-0034762-169; Forward 5′-GAAATGCCCCTTCACTGGTA-3′ and Reverse 5′-TGTGACGATGTCACCGATCT-3′ for hcirc-0051908-208; Forward 5′-TTCTCCCACTCTGGGCTCT-3′ and Reverse 5′-GAGTCTTGGGTCTCCCAGAA-3′ for hcirc-0092283-211; Forward 5′-CCAGGTGGTCTCCTCTGA-3′ and Reverse 5′-GCCAAATCGTTGT-3′ for GAPDH; Forward 5′-AGTGGCAGGGAGGCTGG-3′ and Reverse 5′- GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCTC-5′ for hsa-miR-1207-5p; Forward 5′- GGGGGCAGGAGGGGC-3′ and Reverse 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCTGA-3′ for hsa-miR-328-5; Forward 5′-GGACAGCAGGCACAGACA-3′ and Reverse 5′- GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTGCC-3′ hsa-miR-214-3p; Forward 5′-TGAGGGGCTGGCTTTCC-3′ and Reverse 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGACCAG-3′ for has-miR-185-3P; Forward 5′-CTCGCTTCGGCAGCACA-3′ and Reverse 5′-AACGCTTCACGAATTTGCGT-3′U6; Forward 5′- GGTGGGGACAAGACAGAAGA-3′ and Reverse 5′-CTAAAAGCCCCTTCCCAGAG-3′ for DAB2IP; Forward 5′- CCTGTCTCAGGTGTGAGCAA-3′ and Reverse 5′-GGACTGACCCCACACTCTGT-3′ for nuclear factor I C (NFIC); Forward 5′-TGGAAGGTCAGGGAAACATC-3′ and Reverse 5′-GCTGACTGTGAACTCCCTCC-3′ for IL-27; Forward 5′- CTAAAGAGACCGCGTTCCAG-3′ and Reverse 5′-TGGTGACTGAGGAAGGAAGG-3′ for hypermethylated in cancer 1 (HIC1). For the methylation-specific PCR, the DAB2IP promoter region was amplified by using primers of M-Forward 5′-TTTTTTAATGTTTTTAGTTAGGTTGC-3′ and M-Reverse 5′-CTCCTTTTATATTCCATCTAACGAC-3′.
Luciferase assay
Dual luciferase reporter system psiCHECKTM (Fisher Scientific) was used for luciferase assay. The circRNA-5692 (WT) and its mutant sequences were cloned into the plasmid psiCHECK2. HEK293T cells (4 × 104 cells/well) were cultured in 24-well plates overnight and transfected with 400 ng of psiCHECK vector, psiCHECK-circRNA-5692WT, psiCHECK-circRNA-5692 mutant, psiCHECK-DAB2IP WT, or psiCHECK-DAB2IP mutant, together with the plasmid for Renilla luciferase expression by lipofectamine 2000. One day later, the cells were lysed and their luciferase activities were measured by using Dual-luciferase reporter assay system (Promega). Some luciferase assays were performed after co-transfection with miR-328-5P mimics or its mutant.
Western blotting
The different groups of cells were lysed in lysis buffer supplemented with protease inhibitors (Roche, Indianapolis, USA) and centrifuged. The protein concentrations were determined by a BCA kit (Pierce, Rockford, USA). The cell lysates (10–20 µg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 10–12% gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, USA). After blocking with 5% bovine serum albumin (BSA), the membranes were probed with primary antibodies for 4 h at 37 °C and detected with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000, AP308P, Sigma-Aldrich), followed by visualization by using enhanced chemiluminescent reagents. The primary antibodies included anti-E-cadherin (1:1000, ab1416, Abcam), anti-Vimentin (1:1000, ab137321, Abcam), anti-β-actin (1:1000, ab227387, Abcam), and anti-Snail (1:1000, ab53519, Abcam). The relative levels of each protein expression were determined by densitometric analysis by using ImageJ software.
MTT assay
The proliferation of HCC cells was measured in sextuplicate by MTT assay by using Cell Proliferation Kit (Sigma-Aldrich) according to the manufacturer's instructions.
Flow cytometry
The frequency of apoptotic HCC cells was examined by flow cytometry by using FITC-labeled Annexin V and propidium iodide (PI) in a flow cytometer (Attune NxT, ThermoFisher).
Wound-healing assay
The wound healing of HCC cells was measured. HCC cells were cultured in six-well plates up to ~100% confluence. The cells were starved for 6 h and wounded with a sterile 200-μL pipette tip. After being washed, the cells were cultured for 12 h and photoimaged before and after 12-h culture.
Transwell invasion assay
The invasion of different groups of HCC cells was determined by transwell invasion assay. Briefly, HCC cells (105 cells/well) were cultured in serum-free medium in the top chamber that had been loaded with Matrigel. The bottom chambers were filled with 600 μL of complete medium. After 24–48 h of incubation, the cells on the upper surface of the top chamber were removed with a cotton swab, and the invaded cells on the bottom surface of the top chamber were stained with Harris hematoxylin solution (Sigma) and photoimaged under a light microscope.
Bioinformatics analysis
The potential target miRNAs of circRNA-5692 were predicted by using the tool in bioinformatics database CircNet and further predicted by Shanghai Kangcheng Biotech. The potential target genes of miR-328-5p were predicted by using the miRDB (http://mirdb.org/) and TargetScanHuman (www.targetscan.org).
HCC animal model
C57BL/6 nude mice were obtained from Hunan Slack Jingda Experimental Animal (Changsha, China, the experimental animal production license number: SCXK (Xiang) 2016-0002) and injected with 4 × 106 WT Huh-7 cells (control), the vehicle-transfected Huh-7 cells, or circRNA-5692-overexpressed Huh-7 cells (15 nude mice were randomly divided into 3 groups, 5 in each group). The growth of implanted HCC tumors was monitored for their volumes every 5 days up to 30 days post inoculation. The mice were killed and their xenografts were photoimaged and measured.
Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Comparisons among groups were analyzed by one-way ANOVA and post hoc Tukey tests, and the differences between groups were analyzed by two-tailed t tests by using SPSS 10.0 for Windows. A P-value of < 0.05 was considered statistically significant.
Results
The expression of circRNA-5692 is downregulated in HCC
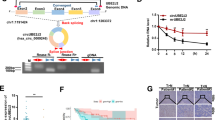
To investigate the potential role of circRNAs in regulating the progression of HCC, 92 pairs of surgical and biopsied HCC and paracarcinoma non-tumor tissues were obtained, and the demographic and clinical charecteristics of those patients are shown in Table 1. The expression profile of circRNAs in HCC and non-tumor liver tissues was analyzed by microarray. In comparison with the non-tumor tissues, there were 103 differentially expressed circRNAs, including 32 upregulated and 71 downregulated in HCC tissues (Fig. 1a). Further analysis of four upregulated and four downregulated circRNAs validated six out of eight differential expression in five pairs of specimens in Fig. 1b, and fiveout of six circRNAs were differentially expressed in HCC Huh-7, SMMC-7721, HCCLM3, MHCC997H, and HepG2 cells (Fig. 1c). It was notable that the circRNA-5692 was encoded by the GLIS2 gene and most significantly downregulated in five HCC tissues and HCC cells (Fig. 1b, c). Further analyses revealed that the relative levels of circRNA-5692 expression in 92 HCC tissues were significantly lower than those in the para-non-tumor liver tissues (P < 0.01, Fig. 1d). Stratification analyses indicated that the lower circRNA-5692 expression was significantly associated with abnormal higher levels of AFP (P = 0.001), cirrhosis history (P = 0.001), larger tumor size (P = 0.042), and distant metastasis (P = 0.025), but not other measures tested in this population. Hence, downregulated circRNA-5692 expression may be associated with the progression of HCC.
The differential expression of circRNAs in three pairs of HCC and non-tumor liver tissues was analyzed by microarray. a The heat mapping of some differentially expressed circRNAs. b Quantitative RT-PCR analysis of relative expression levels of the indicated circRNAs in five pairs of specimens. c Quantitative RT-PCR analysis of relative levels of the indicated circRNAs in different types of cells. d Quantitative RT-PCR analysis of relative levels of has_circ_0005692 (circRNA-5692) in 92 pairs of HCC and non-tumor liver tissues. Data are a representative image or expressed as the mean or mean ± SEM of each group from three separate experiments. **P < 0.01. c cancer tissues, p para-non-tumor tissues.
CircRNA-5692 overexpression suppresses the malignant behaviors of HCC cells
To investigate the potential functions of circRNA-5692 in regulating the progression of HCC, we generated circRNA-5692 stably overexpressing Huh-7 and HepG2 cells by transducing them with lenti-pHBLV-CMV-circRNA-5692 (Fig. 2a). In comparison with the controls, circRNA-5692 overexpression decreased the proliferation, wound healing, and invasion of both Huh-7 and HepG2 cells (P < 0.05, P < 0.01, Fig. 2b–d). Flow cytometry analysis indicated that circRNA-5692 overexpression increased the frequency of spontaneously apoptotic Huh-7 and HepG2 cells (P < 0.01, Fig. 2e). Western blot displayed that circRNA-5692 overexpression significantly increased the relative levels of E-cadherin expression, but decreased Vimentin and Snail expression in both Huh-7 and HepG2 cells. Thus, circRNA-5692 overexpression inhibited the malignant behaviors by attenuating the EMT process in HCC cells (Fig. 2f).
Huh-7 and HepG2 cells were transduced with control lentivirus or virus for expression of circRNA-5692. The proliferation, wound healing, invasion, and apoptosis of HCC cells were measured. The relative levels of EMT-relevant protein expression were determined. a The relative levels of circRNA-5692 expression in HCC cells. b circRNA-5692 overexpression suppressed cell proliferation of Huh-7 and HepG2. c circRNA-5692 overexpression inhibited the wound healing of HCC cells. d circRNA-5692 overexpression attenuated the invasion of HCC cells. e circRNA-5692 overexpression promoted the apoptosis of HCC cells. f Western blotting analysis of the relative levels of the indicated protein expression in different groups of cells. Data are representative images, charts, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. circ-OE. OE overexpression, NC transduced with control lentivirus.
CircRNA-5692 acts as a ceRNA to sponge miRNA-328-5p
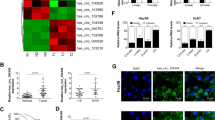
It is well known that a circRNA can act as a ceRNA to sponge miRNAs to mitigate their inhibitory effect on the targeted mRNA expression19. To understand the action of circRNA-5692, we first predicted the potential targeting miRNAs in CircNet database by bioinformatics. As shown in Fig. 3a, circRNA-5692 (purple) was predicted to interact with miR-1207-5p, miR-4763-3p, and miR-4736 (yellow), which would target several gene mRNAs (blue) as well as miR-328-5p, miR-128-1-5p, miR-185-3p, miR-214-3p, and miR-128-2-5p (Fig. 3b). Given that miR-1207-5p, miR-328-5p, miR-185-3p, and miR-214-3p can regulate tumorigenesis20,21,22,23, they were selected for the potential miRNAs targeted by circRNA-5692. Actually, the expression of hsa-miR-1207-5p, hsa-miR-328-5p, and hsa-miR-185-3p increased in five HCC tissues, compared with their para-cancerous tissues (Fig. 3c). However, the expression of hsa-miR-214-3p decreased in four out of five HCC specimens. Similarly, the expression of miRNA-1207-5p and miRNA-328-5p, but not miR-185-3p and miR-214-3p, was upregulated in HCC Huh-7 and HepG2 cells, compared with Chang liver and non-tumor WR168 cells (Fig. 3d). Hence, miRNA-1207-5p and miRNA-328-5p were considered as the miRNA targets of circRNA-5692. Furthermore, luciferase assays indicated that co-transfection with miRNA-328-5p mimics, but not miRNA-328-5p mutant, or miRNA-1207-5p mimics, significantly mitigated the circRNA-5692-regulated luciferase activity in HEK293T cells (P < 0.01, Fig. 3e). Moreover, induction of circRNA-5692 overexpression significantly decreased the relative levels of miRNA-328-5p in both Huh-7 and HepG2 cells (P < 0.01, Fig. 3f). Collectively, these results suggest that circRNA-5692 may sponge miRNA-328-5p in HCC cells.
a The potentially targeted miRNAs of circRNA-5692 were predicted by bioinformatics. b The potential binding sequences of circRNA-5692 to these miRNAs were analyzed by bioinformatics. c Quantitative RT-PCR analysis of the relative RNA levels of the indicated miRNAs in five pairs of HCC tissues. d Quantitative RT-PCR analysis of the relative RNA levels of the indicated miRNAs in the indicated cells. e miR-328-5p overexpression suppressed the circRNA-5692-regulated luciferase activity in HEK293T cells. f circRNA-5692 overexpression decreased miR-328-5p expression in HCC cells. Data are images, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. circ-OE. OE overexpression, NC transduced with control lentivirus. c cancer tissues, p para-non-tumor tissues.
MiRNA-328-5p enhances the malignant behaviors of HCC cells
To investigate the role of miRNA-328-5p in regulating HCC progression, we generated miRNA-328-5p stably overexpressing or silencing Huh-7 and HepG2 cells (Fig. 4a). Compared with the controls, miRNA-328-5p overexpression significantly enhanced the proliferation and wound healing of Huh-7 and HepG2 cells, while miRNA-328-5p silencing attenuated the proliferation and wound healing of Huh-7 and HepG2 cells in vitro (P < 0.01 for all, Fig. 4b, c). Similar patterns of invasion were detected in the different groups of cells (P < 0.01 for all, Fig. 4d). While miRNA-328-5p overexpression significantly decreased the frequency of apoptotic HCC cells, miRNA-328-5p silencing significantly increased it in both Huh-7 and HepG2 cells (P < 0.01 for all, Fig. 4e). Given that the EMT process is associated with cancer cell invasion, we tested the impact of altered miRNA-328-5p expression on the relative levels of E-cadherin, Snail, and Vimentin expression in HCC cells by Western blot (Fig. 4f). The results revealed that miRNA-328-5p overexpression significantly decreased the levels of E-cadherin, but increased Snail and Vimentin expression, while miRNA-328-5p silencing had opposite effects in Huh-7 and HepG2 cells. Such data indicated that miRNA-328-5p enhanced the malignant behaviors of HCC cells by promoting the EMT process.
Huh-7 and HepG2 cells were transfected with control or miR-328-5p mimics or transduced with control lentivirus or lentivirus for expression of miR-328-5p-specific shRNA. The relative levels of miR-328-5p expression in HCC cells were determined with qRT-PCR, and the proliferation and wound healing were examined. a MiR-328-5p expression. b The prolfieration of HCC cells. c The wound healing. d The invasion of HCC cells. e The apoptosis of HCC cells. f Western blot analysis for the relative levels of EMT-relevant protein expression in HCC cells. Data are images, charts, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. miR-OE; ▿▿P < 0.01 vs. miR-sh. OE overexpression, NC transfected with control RNA or lentivirus, Sh transduced with lentivirus for miR-328-5p-specific shRNA.
MiRNA-328-5p targets the DAB2IP expression
Next, the potential target genes of miRNA-328-5p were predicted by bioinformatics by using miRDB (http://mirdb.org/) and TargetScanHuman(www.targetscan.org). There were 336 genes predicted as putative target genes of miRNA-328-5p by miRDB, and 339 by TargetScanHuman (Fig. 5a). Because miRNA-328-5p enhanced the malignant behaviors of HCC cells, we searched the putative target genes with tumor-suppressive function. Among the potential target genes of miRNA-328-5p, the DAB2IP, NFIC, IL-27, and HIC1 genes were tumor suppressors (Fig. 5b). Actually, their mRNA transcripts obviously decreased in five HCC specimens, compared with their para-non-tumor liver tissues (Fig. 5c). Their mRNA transcripts also decreased in the majority of HCC cells tested (Fig. 5d). Further luciferase assays revealed that transfection with miRNA-328-5p mimics, but not its mutant, significantly mitigated the DAB2IP-regulated luciferase activity in HEK293T cells (P < 0.01, Fig. 5e). However, transfection with either miRNA-328-5p mimics or its mutant failed to alter the HIC1, IL-27, or NFIC-regulated luciferase activity in the same experimental conditions. Finally, while miR-328-5p overexpression significantly decreased DAB2IP mRNA transcripts, miR-328-5p silencing dramatically increased DAB2IP mRNA transcripts in Huh-7 and HepG2 cells (Fig. 5f). Such data suggest that miRNA-328-5p may target the DAB2IP mRNA to enhance the malignant behaviors of HCC.
a, b Bioinformatics predicted the targeted mRNAs. c, d Quantitative RT-PCR analysis of the relative levels of the indicated mRNAs in five pairs of HCC tissues and cells. e Luciferase assays revealed that transfection with miR-328-5p mimics inhibited the DAB2IP-controlled luciferase activity in HEK293T cells. f Altered miR-328-5p expression changed the relative levels of DAB2IP mRNA transcripts in HCC cells. Data are images, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. miR-OE; ▿▿P < 0.01 vs. miR-sh. OE overexpression, NC transfected with control RNA or lentivirus, Sh transduced with lentivirus for miR-328-5p-specific shRNA.
DAB2IP attenuates the malignant behaviors of HCC cells
Previous studies have shown that downregulated DAB2IP expression is associated with poor prognosis of HCC24,25. To understand the consequence of miR-328-5p-regulated DAB2IP expression, we generated DAB2IP stably overexpressing and silencing HCC cells (Fig. 6a). Compared with the controls, DAB2IP overexpression significantly inhibited the proliferation, wound healing, and invasion of both Huh-7 and HepG2 cells, and significantly promoted the apoptosis of both Huh-7 and HepG2 cells (P < 0.01 for all, Fig. 6b–e). In contrast, DAB2IP silencing displayed opposite effects on these HCC cells. Further, Western blot indicated that DAB2IP overexpression increased the relative levels of E-cadherin, but decreased Snail and Vimentin expression in Huh-7 and HepG2 cells, while DAB2IP silencing exhibited the reverse effects in Huh-7 and HepG2 cells (Fig. 6f). Thus, DAB2IP, like the circRNA-5692, attenuated the malignant behaviors of HCC cells.
Huh-7 and HepG2 cells were transfected with control lentivirus or lentivirus for DAB2IP expression or DAB2IP-specific shRNA expression. a The relative levels of DAB2IP mRNA transcripts were determined by quantitative RT-PCR. b–e The proliferation, wound healing, invasion, and apoptosis of HCC cells were examined. f The relative levels of EMT-relevant protein expression in the different groups of cells were determined by Western blot. Data are images, charts, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. DAB2IP-OE; ▿▿P < 0.01 vs. DAB2IP-sh. OE overexpression, NC transfected with control RNA or lentivirus, Sh transduced with lentivirus for DAB2IP-specific shRNA.
CircRNA-5692 overexpression attenuates the growth of HCC in vivo
Finally, we tested the impact of circRNA-5692 overexpression on the growth of implanted Huh-7 tumors in vivo. C57BL/6 nude mice were injected with control Huh-7 cells, control lentivirus-transduced Huh-7 cells, or circRNA-5692-overexpressing Huh-7 cells to establish solid tumors. Compared with the control mice, circRNA-5692 overexpression significantly attenuated the growth of implanted Huh-7 tumors (P < 0.01, Fig. 7a) and reduced the tumor size and weights in mice (Fig. 7b, c). Furthermore, circRNA-5692-overexpressing tumors displayed significantly higher levels of circRNA-5692 and DAB2IP mRNA transcripts, but lower miR-328-5p expression (Fig. 7d). Given that high DAB2IP methylation is commonly observed in various tumor tissues, we evaluated the methylation state of DAB2IP in grafted HCC tumors26. By using unmethylated PCR primers, we detected DAB2IP DNA fragments in the circRNA-5692-overexpressing tumors, but not clearly in the control tumors (Fig. 7e), indicating that the DAB2IP methylation was downregulated in the circRNA-5692-overexpressing tumors. Compared with the controls, significantly higher levels of E-cadherin and DAB2IP expression, but lower levels of Snail and Vimentin expression, were detected in the circRNA-5692-overexpressing tumors (Fig. 7f). Therefore, circRNA-5692 overexpression attenuated the growth of implanted Huh-7 tumors in vivo by sponging miR-328-5p to enhance DAB2IP expression.
C57BL/6 nude mice were implanted subcutaneously with Huh-7, Huh-7/NC, or Huh-7/OE cells (n = 3 per group). a The dynamic growth of implanted tumors was monitored longitudinally. b, c The tumor sizes were imaged and their weights were measured. d The relative levels of circRNA-5692, miR-328-5p, and DAB2IP mRNA transcripts in tumor tissues were determined by quantitative RT-PCR. e The methylation status of the DAB2IP promoter of tumor tissues was determined by PCR. f The relative levels of EMT-relevant protein expression in the tumor tissues were determined by Western blot. Data are images, or expressed as the mean ± SEM of each group from three separate experiments. ##P < 0.01 vs. CON; ▵▵P < 0.01 vs. circ-OE. OE overexpression, NC transduced with control lentivirus.
Discussion
Tumor cells usually display malignant behaviors, such as rapid proliferation, migration, invasion, and resistance to apoptosis27. Previous studies have shown that miR-221 and miR-224 can regulate the proliferation and invasion of HCC cells28,29. In this study, we identified differentially expressed circRNAs that were downregulated in clinical HCC tissues, compared with non-tumor liver tissues by microarray. We found that 71 circRNAs were downregulated in HCC tissues, and validated that the randomly selected six circRNAs, particularly for circRNA-5692, decreased their expression in HCC tissues and cells. Further validation in 92 pairs of HCC and non-tumor liver tissues indicated that downregulated circRNA-5692 expression was significantly associated with smaller tumor size, multiple nodules, and higher differentiation of HCC in this population. To the best of our knowledge, this was the first report on downregulated circRNA-5692 expression in HCC, which extended previous reports of other circRNAs30,31,32,33. The downregulated circRNA-5692 expression suggests that circRNA-5692 may act as a tumor suppressor to attenuate the malignant behaviors of HCC. Actually, we found that circRNA-5692 overexpression attenuated the malignant behaviors of HCC cells in vitro and HCC growth in vivo, accompanied by inhibiting the EMT process. Given that the EMT process is crucial for the invasion and metastasis of HCC34,35,36, the reduced EMT process by circRNA-5692 indicates that circRNA-5692 may be an inhibitor of HCC invasion and metastasis. Hence, circRNA-5692 may be a therapeutic target for inhibition of HCC metastasis, and our findings may shed new light on the pathogenesis of HCC.
It is well known that a circRNA can bind to its targeted miRNAs and act as a ceRNA to sponge these miRNAs and inhibit their activity19. While miRNAs bind to the 3′UTR of mRNAs to suppress their translation, and promote their degradation37, circRNA through sponging the miRNAs would enhance the miRNA-targeted gene expression. Actually, circRNA Cdr1as can sponge miR-7 to enhance CCNE1 and PIK3CD gene expression and SMMC-7721 cell proliferation38. In this study, we identified that circRNA-5692 targeted miR-328-5p in HCC cells because miR-328-5p overexpression mitigated the circRNA-5692-regulated luciferase activity, while circRNA-5692 overexpression decreased miR-328-5p expression in HCC cells, accompanied by modulating the EMT process. More importantly, miR-328-5p overexpression enhanced the malignant behaviors, while its silencing attenuated them in HCC cells. These novel data support the notion that miR-328-5p acts as an oncogenic factor to enhance the malignant behaviors of different types of cancers39,40. Therefore, our findings may provide new insights into the molecular regulation of circRNA-569 on the circRNA/miRNA network to regulate the progression of HCC.
We further found that miR-328-5p targeted the DAB2IP, one tumor suppressor, because miR-328-5p expression was inversely associated with DAB2IP expression in HCC tissues and cells, and miR-328-5p overexpression mitigated the DAB2IP-regulated luciferase activity. Furthermore, DAB2IP overexpression attenuated the malignant behaviors and inhibited the EMT process of HCC cells, while DAB2IP silencing displayed opposite effects on HCC cells. Given that DAB2IP is a protein in the RAS–GTPase family26 and acts as a tumor suppressor for different kinds of tumors41, such data indicated that miR-328-5p targeted DAB2IP mRNA to attenuate its expression, together with decreased circRNA-5692 expression to reduce its sponging activity, and promoted the progression of HCC. Therefore, the circRNA-5692/miR-328-5p/DAB2IP pathway may be critical for the development and progression of HCC.
In this study, we failed to detect the DAB2IP promoter region by PCR by using unmethylated primers in the control HCC tumors, consistent with previous observations that hypermethylation of the DAB2IP promoter region is responsible for its downregulated expression in different types of malignant tumors42,43. In contrast, we found that circRNA-5692 overexpression decreased the methylation levels of the DAB2IP promoter region in the HCC xenograft tumors. We understand that one circRNA or miRNA can target several mRNAs, while one mRNA can be targeted by several miRNAs. Furthermore, circRNAs can directly bind to transcription factors and proteins to regulate their functions. The decreased methylation by circRNA-5692 overexpression may stem from the fact that circRNA-5692 may interact with methyltransferase to decrease the methylation levels of DAB2IP promoter region and enhance its expression in HCC. We are interested in further investigating the molecular mechanisms by which circRNA-5692 decreases the methylation of the DAB2IP promoter region in the HCC.
In summary, our data indicated that circRNA-5692 was downregulated in HCC tissues and cells, and acted as a tumor suppressor to attenuate the malignant behaviors of HCC cells, accompanied by inhibiting the EMT process. Furthermore, circRNA-5692 effectively sponged miR-328-5p, which targeted the DAB2IP to enhance the malignant behaviors of HCC cells, while the DAB2IP effectively suppressed the malignant behaviors of HCC cells. Moreover, circRNA-5692 overexpression attenuated the EMT process and implanted HCC tumor growth in vivo by promoting demethylation in the DAB2IP gene. Hence, the circRNA-5692/miR-328-5p/DAB2IP pathway may be critical for regulating the development and progression of HCC and may be a therapeutic target for intervention of HCC. Therefore, our findings may shed new light on the pathogenesis of HCC.
References
European Association for Study of, L., European Organisation for, R. & Treatment of, C. EASL-EORT management of hepatocellular carcinoma. Eur. J. Cancer 48, 599–641 (2012).
Venook, A. P., Papandreou, C., Furuse, J. & de Guevara, L. L. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 15 (Suppl 4), 5–13 (2010).
Sayiner, M., Golabi, P. & Younossi, Z. M. Disease burden of hepatocellular carcinoma: a global perspective. Dig. Dis. Sci. 64, 910–917 (2019).
Oliveri, R. S., Wetterslev, J. & Gluud, C. Hepatocellular carcinoma. Lancet 380, 470–471 (2012). 470; author reply.
Chatterjee, M. & Sengupta, S. Emerging roles of long non-coding RNAs in cancer. J. Biosci. 44, pii: 22 (2019).
Shang, Q., Yang, Z., Jia, R. & Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 18, 6 (2019).
Li, P. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta 444, 132–136 (2015).
Li, P. et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 116, 626–633 (2017).
Chen, J. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 388, 208–219 (2017).
Xia, W. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 6, 35576 (2016).
Li, X., Yang, L. & Chen, L. L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442 (2018).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Li, R. C. et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 9, 838 (2018).
Zhang, X. et al. Circular RNA Vav3 sponges gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol. 16, 118–132 (2019).
Du, W. W. et al. Identifying and characterizing circRNA-protein interaction. Theranostics 7, 4183–4191 (2017).
Du, W. W. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 38, 1402–1412 (2017).
Holdt, L. M. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429 (2016).
Wong, C. M., Tsang, F. H. & Ng, I. O. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 15, 137–151 (2018).
Wang, F., Nazarali, A. J. & Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res 6, 1167–1176 (2016).
Luo, X. et al. MicroRNA-328 enhances cellular motility through posttranscriptional regulation of PTPRJ in human hepatocellular carcinoma. Onco Targets Ther. 8, 3159–3167 (2015).
Wang, C. et al. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J. Gynecol. Oncol. 29, e99 (2018).
Liu, C. et al. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol. Lett. 13, 2631–2636 (2017).
Lu, G. et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res 37, 289 (2018).
Zhang, X. et al. Low expression of DAB2IP contributes to malignant development and poor prognosis in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 27, 1117–1125 (2012).
Qiu, G. H. et al. Differential expression of hDAB2IPA and hDAB2IPB in normal tissues and promoter methylation of hDAB2IPA in hepatocellular carcinoma. J. Hepatol. 46, 655–663 (2007).
Liu, L., Xu, C., Hsieh, J. T., Gong, J. & Xie, D. DAB2IP in cancer. Oncotarget 7, 3766–3776 (2016).
Balogh, J. et al. Hepatocellular carcinoma: a review. J. Hepatocell. Carcinoma 3, 41–53 (2016).
Park, J. K. et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res 71, 7608–7616 (2011).
Ma, D., Tao, X., Gao, F., Fan, C. & Wu, D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol. Lett. 4, 483–488 (2012).
Fu, L., Jiang, Z., Li, T., Hu, Y. & Guo, J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. (2018). https://doi.org/10.1002/cam4.1574. [Epub ahead of print].
Li, M. F. et al. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed. Pharmacother. 99, 848–858 (2018).
Zhang, X. et al. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 11, 2853–2863 (2018).
Zhong, L. et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys. Res Commun. 499, 1044–1049 (2018).
Kota, P. et al. M-Ras/Shoc2 signaling modulates E-cadherin turnover and cell-cell adhesion during collective cell migration. Proc. Natl. Acad. Sci. USA 116, 3536–3545 (2019).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Mendez, M. G., Kojima, S. & Goldman, R. D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 24, 1838–1851 (2010).
Catalanotto, C., Cogoni, C. & Zardo, G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci 17, pii: E1712 (2016).
Yu, L. et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 11, e0158347 (2016).
Zhang, S., Li, P., Zhao, L. & Xu, L. LINC00210 as a miR-328-5p sponge promotes nasopharyngeal carcinoma tumorigenesis by activating NOTCH3 pathway. Biosci Rep 38, pii: BSR20181168 (2018).
Luo, T., Yan, Y., He, Q., Ma, X. & Wang, W. miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by targeting RAGE. Oncol. Rep. 39, 2906–2914 (2018).
Bellazzo, A., Di Minin, G. & Collavin, L. Block one, unleash a hundred. Mechanisms of DAB2IP inactivation in cancer. Cell Death Differ. 24, 15–25 (2017).
Smits, M. et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin. Cancer Res 18, 4048–4058 (2012).
Min, J. et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat. Med 16, 286–294 (2010).
Acknowledgements
This work was supported by grants from International Scientific and Technology Cooperation Program of China (No. 2015DFA31490), the National Natural Sciences Foundation of China (No. 81700561, 81402623), Medical Scientific Research Program of Beijing Medical and Health Foundation (No. YWJKJJHKYJJ-1317459), Hunan Provincial Natural Science Foundation (No. 2019JJ40456), and Guangxi Provincial Natural Science Foundation (No. 2018GXNSFDA281003).
Author contributions
Z.L., J.Q., and X.F. conceived and designed research; Z.L., Y.Y., and Y.K. collected data and conducted research; Z.L. and Z.H. analyzed and interpreted data; Z.L. wrote the initial paper; X.H. and J.Q. revised the paper; Z.L., J.Q., and X.F. had primary responsibility for final content. All authors read and approved the final paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E. Candi
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Z., Yu, Y., Huang, Z. et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis 10, 900 (2019). https://doi.org/10.1038/s41419-019-2089-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-2089-9
This article is cited by
-
EGFR-Activated JAK2/STAT3 Pathway Confers Neuroprotection in Spinal Cord Ischemia–Reperfusion Injury: Evidence from High-Throughput Sequencing and Experimental Models
Molecular Neurobiology (2024)
-
Circular RNA 0016142 Knockdown Induces Ferroptosis in Hepatocellular Carcinoma Cells via Modulation of the MicroRNA-188-3p/Glutathione Peroxidase 4 Axis
Biochemical Genetics (2024)
-
Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance
Cancer and Metastasis Reviews (2024)
-
Circ_0091579 Knockdown Inhibited HCC Proliferation and Glutamine Metabolism Through miR-1270/YAP1 Axis
Biochemical Genetics (2024)
-
Non-coding RNA methylation modifications in hepatocellular carcinoma: interactions and potential implications
Cell Communication and Signaling (2023)