Abstract
Microorganisms colonize various ecological niches in the human habitat, as they do in nature. Predominant forms of multicellular communities called biofilms colonize human tissue surfaces. The gastrointestinal tract is home to a profusion of microorganisms with intertwined, but not identical, lifestyles: as isolated planktonic cells, as biofilms and in biofilm-dispersed form. It is therefore of major importance in understanding homeostatic and altered host–microorganism interactions to consider not only the planktonic lifestyle, but also biofilms and biofilm-dispersed forms. In this Review, we discuss the natural organization of microorganisms at gastrointestinal surfaces, stratification of microbiota taxonomy, biogeographical localization and trans-kingdom interactions occurring within the biofilm habitat. We also discuss existing models used to study biofilms. We assess the contribution of the host–mucosa biofilm relationship to gut homeostasis and to diseases. In addition, we describe how host factors can shape the organization, structure and composition of mucosal biofilms, and how biofilms themselves are implicated in a variety of homeostatic and pathological processes in the gut. Future studies characterizing biofilm nature, physical properties, composition and intrinsic communication could shed new light on gut physiology and lead to potential novel therapeutic options for gastrointestinal diseases.
Key points
-
Bacteria adopt different lifestyles in their natural habitats, from single planktonic cells to biofilm communities.
-
Polymicrobial biofilms naturally grow throughout the gastrointestinal tract, both at the epithelial surface and in the lumen as mucin-attached and food particle-attached colonies.
-
The biofilm lifestyle influences metabolic behaviour of the microbiota but more research is needed to characterize gut biofilm-specific metabolites and their effects on the host response in health and disease.
-
Polymicrobial and trans-kingdom interactions occur in gut biofilms; deciphering the nature of such interactions might improve our current understanding of the homeostatic relationship between the host and its gut microbiota.
-
Abnormal biofilm features are associated with gastrointestinal diseases; characterization of biofilm alterations and cause-to-effect studies are warranted to elucidate their role in pathophysiology.
-
Investigating biogeographical redistribution of biofilms at mucosal surfaces might provide new tools to characterize microbial alterations associated with gastrointestinal diseases and options for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Costerton, J. W., Geesey, G. G. & Cheng, K.-J. How bacteria stick. Sci. Am. 238, 86–95 (1978).
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. & Lappin-Scott, H. M. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995).
Flemming, H. C. & Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019). A review that offers quantitative proof for the dominance of biofilms in natural environments and proposes a comprehensive definition of biofilms.
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
O’Toole, G., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Bjarnsholt, T. et al. The in vivo biofilm. Trends Microbiol. 21, 466–474 (2013).
Li, S., Konstantinov, S. R., Smits, R. & Peppelenbosch, M. P. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol. Med. 23, 18–30 (2017). A review that summarizes the contribution of microbial biofilms to the physiopathology of CRC.
von Rosenvinge, E. C., O’May, G. A., Macfarlane, S., Macfarlane, G. T. & Shirtliff, M. E. Microbial biofilms and gastrointestinal diseases. Pathog. Dis. 67, 25–38 (2013).
Tytgat, H. L. P., Nobrega, F. L., van der Oost, J. & de Vos, W. M. Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 27, 17–25 (2019).
de Vos, W. M. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 1, 15005 (2015).
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999).
Yasuda, K. et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391 (2015).
Zoetendal, E. G. et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Env. Microbiol. 68, 3401–3407 (2002).
Wang, Y. et al. Laser capture microdissection and metagenomic analysis of intact mucosa-associated microbial communities of human colon. Appl. Microbiol. Biotechnol. 88, 1333–1342 (2010).
Nava, G. M., Friedrichsen, H. J. & Stappenbeck, T. S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011). A study revealing the taxonomic specificity of mucosa-associated and lumenal microbiota in the mouse proximal colon.
Pantanella, F., Valenti, P., Natalizi, T., Passeri, D. & Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: pros and cons of the main techniques currently in use. Ann. Ig. 25, 31–42 (2013).
Johnson, C. H. et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21, 891–897 (2015).
Motta, J. P. et al. Iron sequestration in microbiota biofilms as a novel strategy for treating inflammatory bowel disease. Inflamm. Bowel Dis. 24, 1493–1502 (2018).
Couto, N., Schooling, S. R., Dutcher, J. R. & Barber, J. Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J. Proteome Res. 14, 4207–4222 (2015).
Gil, C. et al. Biofilm matrix exoproteins induce a protective immune response against Staphylococcus aureus biofilm infection. Infect. Immun. 82, 1017–1029 (2014).
Muthukrishnan, G. et al. Exoproteome of Staphylococcus aureus reveals putative determinants of nasal carriage. J. Proteome Res. 10, 2064–2078 (2011).
Santos, T. et al. MALDI mass spectrometry imaging and in situ microproteomics of Listeria monocytogenes biofilms. J. Proteom. 187, 152–160 (2018).
Chua, S. L. et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5, 4462 (2014). This study sheds light on the distinct properties of the biofilm-dispersed phenotype in comparison to planktonic and biofilm lifestyles.
Guilhen, C. et al. Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genomics 17, 237 (2016).
Williamson, K. S. et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 (2012).
O’Toole, G. A. & Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304 (1998).
Ceri, H. et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776 (1999).
Sproule-Willoughby, K. M. et al. In vitro anaerobic biofilms of human colonic microbiota. J. Microbiol. Methods 83, 296–301 (2010). This study demonstrates the ability to grow human colon microbiota under its natural polymicrobial biofilm lifestyle using a microtitre-derived model (Calgary biofilm device).
Motta, J. P. et al. Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm. Bowel Dis. 21, 1006–1017 (2015).
Beatty, J. K. et al. Giardia duodenalis induces pathogenic dysbiosis of human intestinal microbiota biofilms. Int. J. Parasitol. 47, 311–326 (2017).
Motta, J. P. et al. Active thrombin produced by the intestinal epithelium controls mucosal biofilms. Nat. Commun. 10, 3224 (2019). This study demonstrates that a host mediator (protease) control the physical properties of gut biofilms and its spatial exclusion from the gut epithelium.
Goeres, D. M. et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat. Protoc. 4, 783–788 (2009).
Lawrence, J. R., Swerhone, G. D. & Neu, T. R. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42, 215–224 (2000).
Macfarlane, S. & Macfarlane, G. T. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Env. Microb. 72, 6204–6211 (2006).
Fehlbaum, S. et al. Design and investigation of polyferms in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the elderly colon. PLoS ONE 10, e0142793 (2015).
Van den Abbeele, P. et al. Arabinoxylans, inulin and Lactobacillus reuteri 1063 repress the adherent-invasive Escherichia coli from mucus in a mucosa-comprising gut model. NPJ Biofilms Microbiomes 2, 16016 (2016).
Van de Wiele, T., Van den Abbeele, P., Ossieur, W., Possemiers, A. & Marzorati, M. in The Impact of Food Bioactives on Health (eds Verhoeckx, K. et al.) 305–317 (Springer, 2015).
Lee, J. H., Kaplan, J. B. & Lee, W. Y. Microfluidic devices for studying growth and detachment of Staphylococcus epidermidis biofilms. Biomed. Microdevices 10, 489–498 (2008).
Yawata, Y., Nguyen, J., Stocker, R. & Rusconi, R. Microfluidic studies of biofilm formation in dynamic environments. J. Bacteriol. 198, 2589–2595 (2016).
Barroso, E., Cueva, C., Peláez, C., Martinez-Cuesta, M. C. & Requena, T. in The Impact of Food Bioactives on Health (eds Verhoeckx, K. et al.) 319–327 (Springer, 2015).
McDonald, J. A. et al. Simulating distal gut mucosal and luminal communities using packed-column biofilm reactors and an in vitro chemostat model. J. Microbiol. Methods 108, 36–44 (2015).
Jalili-Firoozinezhad, S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019). This technologically innovative study demonstrates the co-culture of human intestinal organoids mounted on a chip-based model together with complex anaerobic gut microbiota.
Diard, M. et al. Caenorhabditis elegans as a simple model to study phenotypic and genetic virulence determinants of extraintestinal pathogenic Escherichia coli. Microbes Infect. 9, 214–223 (2007).
Gerbaba, T. K., Gupta, P., Rioux, K., Hansen, D. & Buret, A. G. Giardia duodenalis-induced alterations of commensal bacteria kill Caenorhabditis elegans: a new model to study microbial-microbial interactions in the gut. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G550–G561 (2015).
Purdy, A. E. & Watnick, P. I. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc. Natl Acad. Sci. USA 108, 19737–19742 (2011).
Engel, P., Martinson, V. G. & Moran, N. A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl Acad. Sci. USA 109, 11002–11007 (2012).
Rendueles, O. et al. A new zebrafish model of oro-intestinal pathogen colonization reveals a key role for adhesion in protection by probiotic bacteria. PLoS Pathog. 8, e1002815 (2012).
Lutz, H. L. et al. A simple microbiome in the European common cuttlefish, Sepia officinalis. mSystems 4, e00177–19 (2019).
Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 (2007).
Ha, K. R., Psaltis, A. J., Tan, L. & Wormald, P. J. A sheep model for the study of biofilms in rhinosinusitis. Am. J. Rhinol. 21, 339–345 (2007).
Dohar, J. E. et al. Mucosal biofilm formation on middle-ear mucosa in a nonhuman primate model of chronic suppurative otitis media. Laryngoscope 115, 1469–1472 (2005).
Buret, A., Ward, K. H., Olson, M. E. & Costerton, J. W. An in vivo model to study the pathobiology of infectious biofilms on biomaterial surfaces. J. Biomed. Mater. Res. 25, 865–874 (1991).
Palestrant, D. et al. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct. Pathol. 28, 23–27 (2004).
Bollinger, R. R. et al. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587 (2003). This study suggests a role for host IgA in the formation of gut biofilms and discusses the concept of ‘immune-inclusion/exclusion’ for IgA.
Banwell, J. G., Howard, R., Cooper, D. & Costerton, J. W. Intestinal microbial flora after feeding phytohemagglutinin lectins (Phaseolus vulgaris) to rats. Appl. Env. Microbiol. 50, 68–80 (1985).
Johansson, M. E., Larsson, J. M. & Hansson, G. C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl Acad. Sci. USA 108, 4659–4665 (2011).
Swidsinski, A., Loening-Baucke, V., Lochs, H. & Hale, L. P. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J. Gastroenterol. 11, 1131–1140 (2005).
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389 (2005). Together with Swidsinski et al. (2005), this study demonstrates the use of microscopy approaches to characterize gut biofilms in the healthy and inflamed colon in mice and humans.
Saafan, M. E., Ibrahim, W. S. & Tomoum, M. O. Role of adenoid biofilm in chronic otitis media with effusion in children. Eur. Arch. Otorhinolaryngol. 270, 2417–2425 (2013).
Dejea, C. M. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA 111, 18321–18326 (2014).
Mark Welch, J. L., Hasegawa, Y., McNulty, N. P., Gordon, J. I. & Borisy, G. G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017).
Kotze, S. H. et al. Spontaneous bacterial cell lysis and biofilm formation in the colon of the Cape Dune mole-rat and the laboratory rabbit. Appl. Microbiol. Biotechnol. 90, 1773–1783 (2011).
Frese, S. A. et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 9, e1004057 (2013). This study shows the presence of Lactobacilli biofilms forming at the mucosal surface of the stomach in mice.
Thaiss, C. A. et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12 (2016).
Bergstrom, K. et al. Proximal colon–derived O-glycosylated mucus encapsulates and modulates the microbiota. Science 370, 467–472 (2020).
Smith, H. F. et al. Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. J. Evol. Biol. 22, 1984–1999 (2009).
Tomkovich, S. et al. Human colon mucosal biofilms and murine host communicate via altered mRNA and microRNA expression during cancer. mSystems 5, e00451-19 (2020).
Udden, S. M., Waliullah, S., Harris, M. & Zaki, H. The ex vivo colon organ culture and its use in antimicrobial host defense studies. J. Vis. Exp. https://doi.org/10.3791/55347 (2017).
Johansson, M. E. et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291 (2014).
Birchenough, G. M., Nystrom, E. E., Johansson, M. E. & Hansson, G. C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016).
Zagato, E. et al. Lactobacillus paracasei CBA L74 metabolic products and fermented milk for infant formula have anti-inflammatory activity on dendritic cells in vitro and protective effects against colitis and an enteric pathogen in vivo. PLoS ONE 9, e87615 (2014).
Tsilingiri, K. et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut 61, 1007–1015 (2012).
Hicks, S., Candy, D. C. & Phillips, A. D. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect. Immun. 64, 4751–4760 (1996).
Grassart, A. et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host Microbe 26, 435–444.e4 (2019).
Sidar, B. et al. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip). Lab. Chip 20, 3552–3562 (2019).
Rumbaugh, K. P. & Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 10, 571–586 (2020).
Jefferson, K. K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173 (2004).
Jensen, E. T., Kharazmi, A., Hoiby, N. & Costerton, J. W. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS 100, 727–733 (1992).
Lockhart, J. S. et al. Mixed species biofilms of Fusobacterium necrophorum and Porphyromonas levii impair the oxidative response of bovine neutrophils in vitro. Anaerobe 47, 157–164 (2017).
Kernien, J. F., Johnson, C. J. & Nett, J. E. Conserved inhibition of neutrophil extracellular trap release by clinical Candida albicans biofilms. J. Fungi 3, 49 (2017).
Guilhen, C. et al. Colonization and immune modulation properties of Klebsiella pneumoniae biofilm-dispersed cells. NPJ Biofilms Microbiomes 5, 25 (2019).
Buret, A. G., Motta, J. P., Allain, T., Ferraz, J. & Wallace, J. L. Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron? J. Biomed. Sci. 26, 1 (2019).
Macfarlane, S., Bahrami, B. & Macfarlane, G. T. Mucosal biofilm communities in the human intestinal tract. Adv. Appl. Microbiol. 75, 111–143 (2011).
Macfarlane, S., McBain, A. J. & Macfarlane, G. T. Consequences of biofilm and sessile growth in the large intestine. Adv. Dent. Res. 11, 59–68 (1997). This study is the origin of our perception of the gut microbiota as a biofilm community, and proposes that a biofilm phenotype influences the microbially cooperative metabolism of carbohydrates within the gastrointestinal tract.
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016). A review of the microbial biogeography throughout the gastrointestinal tract and its relevance in health and disease.
Nadell, C. D., Drescher, K. & Foster, K. R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600 (2016).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Lepage, P. et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11, 473–480 (2005).
Charlson, E. S. et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963 (2011).
Segata, N. et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42 (2012).
Gu, S. et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE 8, e74957 (2013).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Ghoul, M. & Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016).
Madsen, J. S. et al. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Env. Microbiol. 18, 2565–2574 (2016).
Donelli, G., Vuotto, C., Cardines, R. & Mastrantonio, P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 65, 318–325 (2012).
Zmora, N. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.e21 (2018).
Suez, J. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018).
Randal Bollinger, R., Barbas, A. S., Bush, E. L., Lin, S. S. & Parker, W. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J. Theor. Biol. 249, 826–831 (2007).
Kamada, N., Chen, G. Y., Inohara, N. & Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690 (2013).
Raffatellu, M. Learning from bacterial competition in the host to develop antimicrobials. Nat. Med. 24, 1097–1103 (2018).
Cassat, J. E. & Skaar, E. P. Iron in infection and immunity. Cell Host Microbe 13, 509–519 (2013).
Da, Re,S. et al. Identification of commensal Escherichia coli genes involved in biofilm resistance to pathogen colonization. PLoS ONE 8, e61628 (2013).
Chudnovskiy, A. et al. Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–456.e14 (2016).
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 (2017).
Pfeiffer, J. K. & Virgin, H. W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872 (2016).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015). This paper describe the microbial biogeography in the colon and stomach of mice, and demonstrates its alteration in the context of a carbohydrate-rich diet.
Thaiss, C. A. et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014).
Liu, C. Y., Dube, P. E., Girish, N., Reddy, A. T. & Polk, D. B. Optical reconstruction of murine colorectal mucosa at cellular resolution. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G721–G735 (2015).
Flemming, H. C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Srinandan, C. S., Elango, M., Gnanadhas, D. P. & Chakravortty, D. Infiltration of matrix-non-producers weakens the Salmonella biofilm and impairs its antimicrobial tolerance and pathogenicity. Front. Microbiol. 6, 1468 (2015).
Jinno, A. & Park, P. W. Role of glycosaminoglycans in infectious disease. Methods Mol. Biol. 1229, 567–585 (2015).
Murofushi, Y. et al. The toll-like receptor family protein RP105/MD1 complex is involved in the immunoregulatory effect of exopolysaccharides from Lactobacillus plantarum N14. Mol. Immunol. 64, 63–75 (2015).
Bylund, J., Burgess, L. A., Cescutti, P., Ernst, R. K. & Speert, D. P. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J. Biol. Chem. 281, 2526–2532 (2006).
Raffatellu, M. et al. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 73, 3367–3374 (2005).
Speziale, P., Pietrocola, G., Foster, T. J. & Geoghegan, J. A. Protein-based biofilm matrices in Staphylococci. Front. Cell Infect. Microbiol. 4, 171 (2014).
Kaplan, J. B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218 (2010).
Passmore, I. J. et al. Mep72, a metzincin protease that is preferentially secreted by biofilms of Pseudomonas aeruginosa. J. Bacteriol. 197, 762–773 (2015).
Purschke, F. G., Hiller, E., Trick, I. & Rupp, S. Flexible survival strategies of Pseudomonas aeruginosa in biofilms result in increased fitness compared with Candida albicans. Mol. Cell Proteom. 11, 1652–1669 (2012).
Wentworth, C. C., Jones, R. M., Kwon, Y. M., Nusrat, A. & Neish, A. S. Commensal-epithelial signaling mediated via formyl peptide receptors. Am. J. Pathol. 177, 2782–2790 (2010).
Sadowska, B., Wieckowska-Szakiel, M., Paszkiewicz, M. & Rozalska, B. The immunomodulatory activity of Staphylococcus aureus products derived from biofilm and planktonic cultures. Arch. Immunol. Ther. Exp. 61, 413–420 (2013).
Brady, R. A., Leid, J. G., Camper, A. K., Costerton, J. W. & Shirtliff, M. E. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect. Immun. 74, 3415–3426 (2006).
Schooling, S. R. & Beveridge, T. J. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957 (2006).
Kulp, A. & Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 (2010).
Ellis, T. N., Leiman, S. A. & Kuehn, M. J. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78, 3822–3831 (2010).
Morris, J. D. et al. Imaging and analysis of Pseudomonas aeruginosa swarming and rhamnolipid production. Appl. Env. Microbiol. 77, 8310–8317 (2011).
Bonnichsen, L. et al. Lipopeptide biosurfactant viscosin enhances dispersal of Pseudomonas fluorescens SBW25 biofilms. Microbiology 161, 2289–2297 (2015).
Periasamy, S. et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl Acad. Sci. USA 109, 1281–1286 (2012).
Das, T., Sehar, S. & Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Env. Microbiol. Rep. 5, 778–786 (2013).
Fuxman Bass, J. I. et al. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J. Immunol. 184, 6386–6395 (2010).
Mulcahy, H., Charron-Mazenod, L. & Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4, e1000213 (2008).
Blenkiron, C. et al. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS ONE 11, e0160440 (2016).
Koeppen, K. et al. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 12, e1005672 (2016).
Mukherjee, S. & Bassler, B. L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019).
Smith, R. S. et al. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-κB and activator protein-2. J. Immunol. 167, 366–374 (2001).
Shiner, E. K. et al. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 8, 1601–1610 (2006).
Tateda, K. et al. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71, 5785–5793 (2003).
Lyte, M. et al. Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun. 232, 682–686 (1997).
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P. & Kaper, J. B. Bacteria-host communication: the language of hormones. Proc. Natl Acad. Sci. USA 100, 8951–8956 (2003). This study reveals that host hormones can activate microbial quorum sensing.
Karavolos, M. H. et al. Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics 9, 458 (2008).
Zaborina, O. et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 3, e35 (2007).
Naresh, R. & Hampson, D. J. Exposure to norepinephrine enhances Brachyspira pilosicoli growth, attraction to mucin and attachment to Caco-2 cells. Microbiology 157, 543–547 (2011).
Korner, M. & Gebbers, J. O. Clinical significance of human intestinal spirochetosis–a morphologic approach. Infection 31, 341–349 (2003).
Freestone, P. P., Sandrini, S. M., Haigh, R. D. & Lyte, M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 16, 55–64 (2008).
McGuckin, M. A., Linden, S. K., Sutton, P. & Florin, T. H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278 (2011).
Tu, Q. V., McGuckin, M. A. & Mendz, G. L. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 57, 795–802 (2008).
Dwivedi, R. et al. L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 101, 575–589 (2016).
Liu, Z. et al. Vibrio cholerae represses polysaccharide synthesis to promote motility in mucosa. Infect. Immun. 83, 1114–1121 (2015).
Kamphuis, J. B. J., Mercier-Bonin, M., Eutamene, H. & Theodorou, V. Mucus organisation is shaped by colonic content; a new view. Sci. Rep. 7, 8527 (2017).
Sonnenburg, E. D. et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Co, J. Y. et al. Mucins trigger dispersal of Pseudomonas aeruginosa biofilms. NPJ Biofilms Microbiomes 4, 23 (2018).
Batoni, G., Maisetta, G. & Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 1858, 1044–1060 (2016).
de la Fuente-Nunez, C. et al. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205 (2015).
Segev-Zarko, L., Saar-Dover, R., Brumfeld, V., Mangoni, M. L. & Shai, Y. Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem. J. 468, 259–270 (2015).
de la Fuente-Nunez, C. et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704 (2012).
Overhage, J. et al. Human host defence peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182 (2008).
Chu, H. et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 (2012).
Schroeder, B. O. et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469, 419–423 (2011).
Wallace, J. L. Physiological and pathophysiological roles of hydrogen sulfide in the gastrointestinal tract. Antioxid. Redox Signal. 12, 1125–1133 (2010).
Goubern, M., Andriamihaja, M., Nubel, T., Blachier, F. & Bouillaud, F. Sulfide, the first inorganic substrate for human cells. FASEB J. 21, 1699–1706 (2007).
Rowan, F. E., Docherty, N. G., Coffey, J. C. & O’Connell, P. R. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br. J. Surg. 96, 151–158 (2009).
Cai, W. J., Wang, M. J., Ju, L. H., Wang, C. & Zhu, Y. C. Hydrogen sulfide induces human colon cancer cell proliferation: role of Akt, ERK and p21. Cell Biol. Int. 34, 565–572 (2010).
Ankri, S. & Mirelman, D. Antimicrobial properties of allicin from garlic. Microbes Infect. 1, 125–129 (1999).
Ross, Z. M., O’Gara, E. A., Hill, D. J., Sleightholme, H. V. & Maslin, D. J. Antimicrobial properties of garlic oil against human enteric bacteria: evaluation of methodologies and comparisons with garlic oil sulfides and garlic powder. Appl. Env. Microbiol. 67, 475–480 (2001).
Pabst, O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 12, 821–832 (2012).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014).
Levites, Y. et al. A human monoclonal IgG that binds Aβ assemblies and diverse amyloids exhibits anti-amyloid activities in vitro and in vivo. J. Neurosci. 35, 6265–6276 (2015).
Tursi, S. A. et al. Salmonella typhimurium biofilm disruption by a human antibody that binds a pan-amyloid epitope on curli. Nat. Commun. 11, 1007 (2020). This study demonstrates the ability of human immunoglobulins to alter the formation of deleterious biofilms on medical implants.
Barnhart, M. M. & Chapman, M. R. Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 (2006).
Wold, A. E. et al. Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect. Immun. 58, 3073–3077 (1990).
Boren, T., Falk, P., Roth, K. A., Larson, G. & Normark, S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262, 1892–1895 (1993).
Moshier, A., Reddy, M. S. & Scannapieco, F. A. Role of type 1 fimbriae in the adhesion of Escherichia coli to salivary mucin and secretory immunoglobulin A. Curr. Microbiol. 33, 200–208 (1996).
Harris, L. G., Nigam, Y., Sawyer, J., Mack, D. & Pritchard, D. I. Lucilia sericata chymotrypsin disrupts protein adhesin-mediated staphylococcal biofilm formation. Appl. Env. Microb. 79, 1393–1395 (2013).
Xu, W. et al. Host and bacterial proteases influence biofilm formation and virulence in a murine model of enterococcal catheter-associated urinary tract infection. NPJ Biofilms Microbiomes 3, 28 (2017). This study shows that a host protease can act as a defence mechanism against deleterious biofilm growth in mouse bladder.
Iwase, T. et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010).
Krishnaswamy, V. R., Mintz, D. & Sagi, I. Matrix metalloproteinases: the sculptors of chronic cutaneous wounds. Biochim. Biophys. Acta Mol. Cell Res. 1864, 2220–2227 (2017).
Selan, L. et al. Serratiopeptidase: a well-known metalloprotease with a new non-proteolytic activity against S. aureus biofilm. BMC Microbiol. 15, 207 (2015).
Tseng, B. S. et al. A biofilm matrix-associated protease inhibitor protects Pseudomonas aeruginosa from proteolytic attack. mBio 9, 2 (2018).
Eggers, C. T., Murray, I. A., Delmar, V. A., Day, A. G. & Craik, C. S. The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase. Biochem. J. 379, 107–118 (2004).
Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 65, 1215–1224 (2016).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). A review of the different strategies used for biofilm eradication and control for human health purposes.
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P. & Hall-Stoodley, L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755 (2017).
Hathroubi, S., Servetas, S. L., Windham, I., Merrell, D. S. & Ottemann, K. M. Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol. Mol. Biol. Rev. 82, e00001-18 (2018).
Koteish, A., Kannangai, R., Abraham, S. C. & Torbenson, M. Colonic spirochetosis in children and adults. Am. J. Clin. Pathol. 120, 828–832 (2003).
Kaakoush, N. O., Castano-Rodriguez, N., Man, S. M. & Mitchell, H. M. Is Campylobacter to esophageal adenocarcinoma as Helicobacter is to gastric adenocarcinoma? Trends Microbiol. 23, 455–462 (2015).
Erickson, L. A. & Torbenson, M. S. Intestinal spirochetosis. Mayo Clin. Proc. 95, 427–428 (2020).
Donskey, C. J. et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N. Engl. J. Med. 343, 1925–1932 (2000).
Smits, W. K., Lyras, D., Lacy, D. B., Wilcox, M. H. & Kuijper, E. J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2, 16020 (2016).
Yonezawa, H. et al. Assessment of in vitro biofilm formation by Helicobacter pylori. J. Gastroenterol. Hepatol. 25, S90–S94 (2010).
Joshua, G. W., Guthrie-Irons, C., Karlyshev, A. V. & Wren, B. W. Biofilm formation in Campylobacter jejuni. Microbiology 152, 387–396 (2006).
Ethapa, T. et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555 (2013).
Heikens, E., Bonten, M. J. & Willems, R. J. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 189, 8233–8240 (2007).
Dubois, T. et al. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ Biofilms Microbiomes 5, 14 (2019).
Grimm, I., Dumke, J., Dreier, J., Knabbe, C. & Vollmer, T. Biofilm formation and transcriptome analysis of Streptococcus gallolyticus subsp. gallolyticus in response to lysozyme. PLoS ONE 13, e0191705 (2018).
Tan, S., Noto, J. M., Romero-Gallo, J., Peek, R. M. Jr. & Amieva, M. R. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 7, e1002050 (2011).
Sigal, M. et al. Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology 148, 1392–1404.e21 (2015). This study demonstrates the presence of H. pylori biofilms in the gastric glands of mice and their contribution to the pathophysiology of cancer.
He, Z. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68, 289–300 (2019).
Warren, R. L. et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 1, 16 (2013).
Stahl, M. et al. A novel mouse model of Campylobacter jejuni gastroenteritis reveals key pro-inflammatory and tissue protective roles for Toll-like receptor signaling during infection. PLoS Pathog. 10, e1004264 (2014).
Semenyuk, E. G. et al. Analysis of bacterial communities during Clostridium difficile infection in the mouse. Infect. Immun. 83, 4383–4391 (2015).
Soavelomandroso, A. P. et al. Biofilm structures in a mono-associated mouse model of Clostridium difficile infection. Front. Microbiol. 8, 2086 (2017).
Coticchia, J. M. et al. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J. Gastrointest. Surg. 10, 883–889 (2006).
Walker, M. M. et al. Colonic spirochetosis is associated with colonic eosinophilia and irritable bowel syndrome in a general population in Sweden. Hum. Pathol. 46, 277–283 (2015).
Goodsall, T. M. et al. Unique pathology of colonic spirochaetosis characterised by mucosal eosinophilia is linked to diarrhoea and IBS. Gut 66, 978–979 (2017).
Kalischuk, L. D. & Buret, A. G. A role for campylobacter jejuni-induced enteritis in inflammatory bowel disease? Am. J. Physiol. Gastrointest. Liver Physiol 298, G1–G9 (2010).
Spiller, R. & Garsed, K. Postinfectious irritable bowel syndrome. Gastroenterology 136, 1979–1988 (2009).
Castano-Rodriguez, N., Kaakoush, N. O., Lee, W. S. & Mitchell, H. M. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut 66, 235–249 (2017).
Kumar, R. et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 13, e1006440 (2017).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Tjalsma, H., Boleij, A., Marchesi, J. R. & Dutilh, B. E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582 (2012).
Drewes, J. L. et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34 (2017).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013).
Hajishengallis, G., Darveau, R. P. & Curtis, M. A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725 (2012).
Tomkovich, S. et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest. 129, 1699–1712 (2019).
Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 421 (2018).
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012).
Swidsinski, A. et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 122, 44–54 (2002).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Swidsinski, A., Loening-Baucke, V., Vaneechoutte, M. & Doerffel, Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 14, 147–161 (2008).
Glasser, A. L. et al. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69, 5529–5537 (2001).
Wang, M., Molin, G., Ahrne, S., Adawi, D. & Jeppsson, B. High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16S rRNA genes. Dig. Dis. Sci. 52, 620–627 (2007).
Palmela, C. et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587 (2018).
Macfarlane, S., Furrie, E., Cummings, J. H. & Macfarlane, G. T. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38, 1690–1699 (2004).
Golinska, E. et al. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J. Gastroenterol. 19, 3562–3572 (2013).
Martinez-Medina, M. et al. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9, 202 (2009).
Png, C. W. et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428 (2010).
Hickey, C. A. et al. Colitogenic bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe 17, 672–680 (2015).
Gibold, L. et al. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell Microbiol. 18, 617–631 (2016).
Lidell, M. E., Moncada, D. M., Chadee, K. & Hansson, G. C. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc. Natl Acad. Sci. USA 103, 9298–9303 (2006).
van der Post, S. et al. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB). J. Biol. Chem. 288, 14636–14646 (2013).
Kerckhoffs, A. P. M. et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J. Med. Microbiol. 60, 236–245 (2011).
Caminero, A. et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 151, 670–683 (2016).
Reti, K. L., Tymensen, L. D., Davis, S. P., Amrein, M. W. & Buret, A. G. Campylobacter jejuni increases flagellar expression and adhesion of noninvasive Escherichia coli: effects on enterocytic Toll-like receptor 4 and CXCL-8 expression. Infect. Immun. 83, 4571–4581 (2015).
Yuki, N. et al. Colonization of the stratified squamous epithelium of the nonsecreting area of horse stomach by lactobacilli. Appl. Env. Microbiol. 66, 5030–5034 (2000).
Savage, D. C. & Blumershine, R. V. Surface-surface associations in microbial communities populating epithelial habitats in the murine gastrointestinal ecosystem: scanning electron microscopy. Infect. Immun. 10, 240–250 (1974).
Wang, Z. H. et al. Bacterial biofilm bioinspired persistent luminescence nanoparticles with gut-oriented drug delivery for colorectal cancer imaging and chemotherapy. ACS Appl. Mater. Interfaces 11, 40 (2019). This study suggests the use of the biofilm-forming capacity of gut microorganisms to locally deliver a therapeutic molecule of medical interest.
Suez, J., Zmora, N., Segal, E. & Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729 (2019).
Alav, I., Sutton, J. M. & Rahman, K. M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73, 2003–2020 (2018).
Ramiro, R. S., Durao, P., Bank, C. & Gordo, I. Low mutational load and high mutation rate variation in gut commensal bacteria. PLoS Biol. 18, e3000617 (2020).
Oliver, A., Canton, R., Campo, P., Baquero, F. & Blazquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254 (2000).
Sorensen, S. J., Bailey, M., Hansen, L. H., Kroer, N. & Wuertz, S. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710 (2005).
Donlan, R. M. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 17, 66–72 (2009).
Vinodkumar, C. S., Neelagund, Y. F. & Kalsurmath, S. Bacteriophage in the treatment of experimental septicemic mice from a clinical isolate of multidrug resistant Klebsiella pneumoniae. J. Commun. Dis. 37, 18–29 (2005).
Biswas, B. et al. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70, 204–210 (2002).
Lu, T. K. & Collins, J. J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA 104, 11197–11202 (2007).
Singh, R., Sahore, S., Kaur, P., Rani, A. & Ray, P. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog. Dis. 74, ftw056 (2016).
Booijink, C. C. et al. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl. Env. Microbiol. 76, 5533–5540 (2010).
Wan, N. et al. Bacterial metabolism during biofilm growth investigated by (13)C tracing. Front. Microbiol. 9, 2657 (2018).
Heffernan, B., Murphy, C. D. & Casey, E. Comparison of planktonic and biofilm cultures of Pseudomonas fluorescens DSM 8341 cells grown on fluoroacetate. Appl. Env. Microbiol. 75, 2899–2907 (2009).
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. & Davies, D. G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 (2002).
Floyd, K. A. et al. Adhesive fiber stratification in uropathogenic Escherichia coli biofilms unveils oxygen-mediated control of type 1 pili. PLoS Pathog. 11, e1004697 (2015).
Khan, M. T. et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 6, 1578–1585 (2012).
Cummings, J. H. & Macfarlane, G. T. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459 (1991).
Chambers, E. S., Preston, T., Frost, G. & Morrison, D. J. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr. Nutr. Rep. 7, 198–206 (2018).
Rios-Covian, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016).
Bach Knudsen, K. E., Jensen, B. B., Andersen, J. O. & Hansen, I. Gastrointestinal implications in pigs of wheat and oat fractions. 2. Microbial activity in the gastrointestinal tract. Br. J. Nutr. 65, 233–248 (1991).
Suzuki, I., Shimizu, T. & Senpuku, H. Role of SCFAs for fimbrillin-dependent biofilm formation of Actinomyces oris. Microorganisms 6, 114 (2018).
Yoneda, S. et al. Effects of short-chain fatty acids on Actinomyces naeslundii biofilm formation. Mol. Oral Microbiol. 28, 354–365 (2013).
Davies, D. G. & Marques, C. N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 (2009).
Chen, T. et al. Efficient biofilm-based fermentation strategies for l-threonine production by Escherichia coli. Front. Microbiol. 10, 1773 (2019).
Mafra, D., Barros, A. F. & Fouque, D. Dietary protein metabolism by gut microbiota and its consequences for chronic kidney disease patients. Future Microbiol. 8, 1317–1323 (2013).
Mirvish, S. S. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 93, 17–48 (1995).
Pitcher, M. C., Beatty, E. R. & Cummings, J. H. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46, 64–72 (2000).
Attene-Ramos, M. S., Wagner, E. D., Gaskins, H. R. & Plewa, M. J. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 5, 455–459 (2007).
Wallace, J. L., Motta, J. P. & Buret, A. G. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G143–G149 (2018).
Warren, M. J., Raux, E., Schubert, H. L. & Escalante-Semerena, J. C. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19, 390–412 (2002).
Crespo, A., Blanco-Cabra, N. & Torrents, E. Aerobic vitamin B12 biosynthesis is essential for Pseudomonas aeruginosa class II ribonucleotide reductase activity during planktonic and biofilm growth. Front. Microbiol. 9, 986 (2018).
Crespo, A., Pedraz, L., Astola, J. & Torrents, E. Pseudomonas aeruginosa exhibits deficient biofilm formation in the absence of class II and III ribonucleotide reductases due to hindered anaerobic growth. Front. Microbiol. 7, 688 (2016).
Ward, M. G. et al. Prevalence and risk factors for functional vitamin B12 deficiency in patients with Crohn’s disease. Inflamm. Bowel Dis. 21, 2839–2847 (2015).
Hooper, C. A., Haney, B. B. & Stone, H. H. Gastrointestinal bleeding due to vitamin K deficiency in patients on parenteral cefamandole. Lancet 1, 39–40 (1980).
Mahdinia, E., Demirci, A. & Berenjian, A. Strain and plastic composite support (PCS) selection for vitamin K (menaquinone-7) production in biofilm reactors. Bioprocess. Biosyst. Eng. 40, 1507–1517 (2017).
Mahdinia, E., Demirci, A. & Berenjian, A. Optimization of Bacillus subtilis natto growth parameters in glycerol-based medium for vitamin K (menaquinone-7) production in biofilm reactors. Bioprocess. Biosyst. Eng. 41, 195–204 (2018).
Roberfroid, M. B. Prebiotics and probiotics: are they functional foods? Am. J. Clin. Nutr. 71, 1682S–1687S (2000).
Takeno, S. & Sakai, T. Involvement of the intestinal microflora in nitrazepam-induced teratogenicity in rats and its relationship to nitroreduction. Teratology 44, 209–214 (1991).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363, eaat9931 (2019). A comprehensive review of the current knowledge and future directions regarding the role of gut microorganisms in the metabolism of human drugs.
Sousa, T. et al. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 363, 1–25 (2008).
Matuskova, Z. et al. Administration of a probiotic can change drug pharmacokinetics: effect of E. coli Nissle 1917 on amidarone absorption in rats. PLoS ONE 9, e87150 (2014).
Guo, Y. et al. Commensal gut bacteria convert the immunosuppressant tacrolimus to less potent metabolites. Drug Metab. Dispos. 47, 194–202 (2019).
Koppel, N., Bisanz, J. E., Pandelia, M. E., Turnbaugh, P. J. & Balskus, E. P. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife 7, e33953 (2018).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009).
Jia, W., Li, H., Zhao, L. & Nicholson, J. K. Gut microbiota: a potential new territory for drug targeting. Nat. Rev. Drug Discov. 7, 123–129 (2008).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Author information
Authors and Affiliations
Contributions
J.-P.M. and N.V. researched data for the article, made substantial contributions to discussion of content, wrote the article, and reviewed/edited the manuscript before submission. C.D. made a substantial contribution to discussion of content, and reviewed/edited the manuscript before submission. J.L.W. and A.G.B. reviewed/edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
J.L.W. is Chief Science Officer of Antibe Therapeutics. J.-P.M. received salary support from Antibe Therapeutics and from CVasThera. The other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks G. Hold, R. Briandet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Biofilm
-
A microbial lifestyle in which microorganisms are embedded in a biopolymer matrix, attached to surfaces, engaged in collective behaviour (for example, communication, cooperation, competition and differentiation) and able to persist even in hostile environments.
- Planktonic
-
Free-swimming, free-floating, single-cell mode of life of microorganisms.
- Biofilm matrix
-
The biopolymer substance containing communities of microorganisms assembled as a biofilm. The chemical complexity of the matrix is not fully appreciated for in vivo biofilms.
- Polymicrobial
-
A microbial community that harbours diversity in terms of species and/or strain content.
- Inflammatory bowel disease
-
(IBD). Chronic inflammatory disorder of the gastrointestinal tract, with multifactorial aetiology that includes genetic susceptibility and environmental factors. Associated with a displacement of gut ecology and an uncontrolled activation of the immune system.
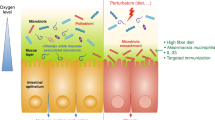
- Oxygen tension
-
Variable oxygenation profile over the intestinal landscape, affecting host immunity and gut microbiota.
- Mucins
-
A class of epithelial gel-forming and non-gel-forming proteins that confer to mucus its viscous hydrophobic property, making it a physical barrier to microorganisms.
- Microbial biogeography
-
Spatial organization of microbial taxa at mucosal surfaces.
- Biofilm-dispersed bacteria
-
Microorganisms that naturally disperse from a biofilm, thereby acquiring biological characteristics distinct from those of their planktonic or biofilm counterparts.
- Commensal
-
A microorganism within the digestive tract that resides in a neutral or beneficial relationship with the host.
- Pathobionts
-
Potentially disease-causing commensals that otherwise (in healthy circumstances) live as non-harmful microorganisms.
- Microbiota stability
-
Reflects the ability of the microbiota to resist environmental stressor-associated perturbations.
- Microbiota resilience
-
Reflects the ability of the microbiota to recover after environmental stressor-associated perturbations.
- Probiotic
-
Microorganisms providing health benefits through direct or indirect effects on intestinal pathobionts, pathogens or host cells.
- Dysbiosis
-
A term used to describe taxonomic, metabolic or structural imbalances that characterize the microbiota associated with a disease condition.
- Substratum
-
The biotic or abiotic surface on which a biofilm can form.
- Quorum sensing
-
Density-dependent cell–cell communication, in which a signal informs the community of a threshold concentration and triggers collective group behaviour and biofilm formation.
- Keystone pathogen
-
A low-abundance pathogen that can trigger a disproportionate effect on tissue by provoking microbiota dysbiosis.
- Gut biofilm biomarkers
-
A molecule, gene or other biological characteristic of gut biofilms that might be used to predict a specific intestinal disease status.
- Xenobiotics
-
Chemical substances that are not naturally produced within the organism.
Rights and permissions
About this article
Cite this article
Motta, JP., Wallace, J.L., Buret, A.G. et al. Gastrointestinal biofilms in health and disease. Nat Rev Gastroenterol Hepatol 18, 314–334 (2021). https://doi.org/10.1038/s41575-020-00397-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-020-00397-y
This article is cited by
-
Autoinducer-2 promotes the colonization of Lactobacillus rhamnosus GG to improve the intestinal barrier function in a neonatal mouse model of antibiotic-induced intestinal dysbiosis
Journal of Translational Medicine (2024)
-
The gut microbiota and its biogeography
Nature Reviews Microbiology (2024)
-
Hypoxia-inducible factor-driven glycolytic adaptations in host-microbe interactions
Pflügers Archiv - European Journal of Physiology (2024)
-
Remedial Measures for Neurodegenerative Diseases Targeting Gut-Microbial Dysfunction with Herbal Bio-Actives
Proceedings of the National Academy of Sciences, India Section B: Biological Sciences (2024)
-
The gut microbiome-prostate cancer crosstalk is modulated by dietary polyunsaturated long-chain fatty acids
Nature Communications (2024)