Abstract
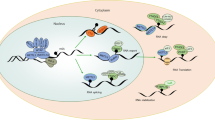
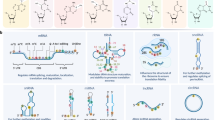
Protection from harmful pathogens depends on activation of the immune system, which relies on tight regulation of gene expression. Recently, the RNA modification N6-methyladenosine (m6A) has been found to play an essential role in such regulation. Here, we summarize newly discovered functions of m6A in controlling various aspects of immunity, including immune recognition, activation of innate and adaptive immune responses, and cell fate decisions. We then discuss some of the current challenges in the field and describe future directions for uncovering the immunological functions of m6A and its mechanisms of action.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Li, S. & Mason, C. E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 15, 127–150 (2014).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Perry, R. P. & Kelley, D. E. Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42 (1974).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Śledź, P. & Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. Elife 5, e18434 (2016).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016); erratum 542, 260 (2017).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ping, X.-L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6 (2018).
Yue, Y. et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018).
Knuckles, P., Lence, T. & Haussmann, I. U. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Garcia-Campos, M. A. et al. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019).
Zhang, Z. et al. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5, eaax0250 (2019).
Darnell, R. B., Ke, S. & Darnell, J. E. Jr. Pre-mRNA processing includes N6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics”. RNA 24, 262–267 (2018).
Gokhale, N. S. et al. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016).
Lichinchi, G. et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20, 666–673 (2016).
Lu, M. et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598 (2020).
Pendleton, K. E. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14 (2017).
Ma, H. et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94 (2019); erratum 15, 549 (2019).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733 (2019).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science 363, eaav0080 (2019).
Boulias, K. et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75, 631–643.e8 (2019).
Sun, H., Zhang, M., Li, K., Bai, D. & Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82 (2019).
Li, F., Zhao, D., Wu, J. & Shi, Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 24, 1490–1492 (2014).
Zhu, T. et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24, 1493–1496 (2014).
Xu, C. et al. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015).
Xu, C. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014).
Nayler, O., Hartmann, A. M. & Stamm, S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 150, 949–962 (2000).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Wojtas, M. N. et al. Regulation of m6A transcripts by the 3′→ 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68, 374–387.e12 (2017).
Bailey, A. S. et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6, 26116 (2017).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019).
Kasowitz, S. D. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14, e1007412 (2018).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6, e31311 (2017).
Kretschmer, J. et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA 24, 1339–1350 (2018).
Hsu, P. J. et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017).
Jain, D. et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7, e30919 (2018).
Arguello, A. E., DeLiberto, A. N. & Kleiner, R. E. RNA chemical proteomics reveals the N6-methyladenosine (m6A)-regulated protein–RNA interactome. J. Am. Chem. Soc. 139, 17249–17252 (2017).
Edupuganti, R. R. et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Alarcón, C. R. et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Spitale, R. C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015).
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Wei, J. et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5 (2018).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Peng, S. et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11, eaau7116 (2019).
Wang, L., Wen, M. & Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365, eaav0758 (2019).
Sommer, S., Lavi, U. & Darnell, J. E. Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499 (1978).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Li, H.-B. et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Lee, H. et al. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell Biol. 21, 700–709 (2019).
Cheng, Y. et al. m6A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 28, 1703–1716.e6 (2019).
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205.e9 (2018).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Zhou, J. et al. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell 69, 636–647.e7 (2018).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Haussmann, I. U. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016).
Lesbirel, S. et al. The m6A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 8, 13827 (2018).
Grozhik, A. V., Linder, B., Olarerin-George, A. O. & Jaffrey, S. R. Mapping m6A at individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP). Methods Mol. Biol. 1562, 55–78 (2017).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Ke, S., Alemu, E. A., Mertens, C. & Gantman, E. C. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes 29, 2037–2053 (2015).
Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280 (2019).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Lorenz, D. A., Sathe, S., Einstein, J. M. & Yeo, G. W. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base specific resolution. RNA 26, 19–28 (2019).
Wu, J. & Chen, Z. J. Innate immune sensing and signaling of cytosolic nucleic acids. Ann. Rev. Immunol. 32, 461–488 (2014).
Roers, A., Hiller, B. & Hornung, V. Recognition of endogenous nucleic acids by the innate immune system. Immunity 44, 739–754 (2016).
Karikó, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Durbin, A. F., Wang, C., Marcotrigiano, J. & Gehrke, L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. Mbio 7, e00833–16 (2016).
Wilusz, J. E. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA 9, e1478 (2018).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e4 (2019).
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5 (2017).
Chen, Y. G. et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9 (2019).
Imaeda, A. et al. N6-methyl adenosine in siRNA evades immune response without reducing RNAi activity. Nucleosides Nucleotides Nucleic Acids 38, 972–979 (2019).
Ringeard, M., Marchand, V., Decroly, E., Motorin, Y. & Bennasser, Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504 (2019).
Habjan, M. et al. Sequestration by IFIT1 impairs translation of 2′ O-unmethylated capped RNA. PLoS Pathog. 9, e1003663 (2013).
Hyde, J. L. & Diamond, M. S. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 479–480, 66–74 (2015).
Daffis, S. et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456 (2010).
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Zheng, Q., Hou, J., Zhou, Y., Li, Z. & Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103 (2017); erratum 18, 1361 (2017).
Winkler, R. et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182 (2019); erratum 20, 243 (2019).
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L. & Mohr, I. RNA m6 A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 32, 1472–1484 (2018).
Zhang, Y. et al. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl Acad. Sci. USA 116, 976–981 (2019).
Liu, Y. et al. N6-methyladenosine RNA modification–mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176 (2019).
Williams, G. D., Gokhale, N. S. & Horner, S. M. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu. Rev. Virol. 6, 235–253 (2019).
Price, A. M., Hayer, K. E., McIntyre, A. B. R. & Gokhale, N. S. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Preprint at bioRxiv https://doi.org/10.1101/865485 (2019).
Gao, Y. et al. Mettl3 mediated m6A modification is essential in fetal hematopoiesis. Blood 132 (Suppl. 1), 3825 (2018).
Wang, H. et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 10, 1898 (2019).
Tan, J. T. et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl Acad. Sci. USA 98, 8732–8737 (2001).
Palmer, D. C. & Restifo, N. P. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30, 592–602 (2009).
Seki, Y.-I. et al. IL-7/STAT5 cytokine signaling pathway is essential but insufficient for maintenance of naive CD4 T cell survival in peripheral lymphoid organs. J. Immunol. 178, 262–270 (2007).
Bahrami, S. & Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 62, 37–49 (2016).
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 (2011).
Sprent, J. & Surh, C. D. Writer’s block: preventing m6A mRNA methylation promotes T cell naivety. Immunol. Cell Biol. 95, 855–856 (2017).
Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 153, 1151–1164 (1995).
Lio, C.-W. J. & Hsieh, C.-S. A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008).
Chinen, T. et al. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 17, 1322–1333 (2016).
Tong, J. et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. 28, 253–256 (2018).
Lu, L.-F. et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30, 80–91 (2009).
Qi, H., Kastenmüller, W. & Germain, R. N. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu. Rev. Cell Dev. Biol. 30, 141–167 (2014).
Dustin, M. L. The immunological synapse. Cancer Immunol. Res. 2, 1023–1033 (2014).
Hawiger, D. et al. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J. Exp. Med. 194, 769–779 (2001).
Liu, K. & Nussenzweig, M. C. Origin and development of dendritic cells. Immunol. Rev. 234, 45–54 (2010).
Feng, Z., Li, Q., Meng, R., Yi, B. & Xu, Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 22, 2558–2568 (2018).
Han, D. et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274 (2019).
Shakhar, G. et al. Stable T cell–dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat. Immunol. 6, 707–714 (2005).
Yamamoto, M. et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420, 324–329 (2002).
Kurts, C., Robinson, B. W. S. & Knolle, P. A. Cross-priming in health and disease. Nat. Rev. Immunol. 10, 403–414 (2010).
Alloatti, A., Kotsias, F., Magalhaes, J. G. & Amigorena, S. Dendritic cell maturation and cross-presentation: timing matters! Immunol. Rev. 272, 97–108 (2016).
Cebrian, I. et al. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 (2011).
Burbage, M., Gros, M. & Amigorena, S. Translate less, prime better, to improve anti-tumor responses. Nat. Immunol. 20, 518–520 (2019).
Li, L. & Clevers, H. Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 (2010).
Kissa, K. & Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010).
Boisset, J.-C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010).
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010).
Zhang, C. et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017).
Lv, J. et al. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 28, 249–252 (2018).
Zhang, P. et al. G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res. 25, 1093–1107 (2015).
Lizama, C. O. et al. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat. Commun. 6, 7739 (2015).
Gama-Norton, L. et al. Notch signal strength controls cell fate in the haemogenic endothelium. Nat. Commun. 6, 8510 (2015).
Yao, Q. J. et al. Mettl3–Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 28, 952–954 (2018).
Reavie, L. et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase–substrate complex. Nat. Immunol. 11, 207–215 (2010).
Wilson, A. et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18, 2747–2763 (2004).
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Wang, H. et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 28, 1035–1038 (2018).
Li, Z. et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28, 904–917 (2018).
Ebert, M. S. & Sharp, P. A. Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012).
Labi, V. et al. Context-specific regulation of cell survival by a miRNA-controlled BIM rheostat. Genes Dev. 33, 1673–1687 (2019).
Acknowledgements
We thank Igor Ulitsky, Schraga Schwartz, Michal Schwartz and Amalie Grenov for providing valuable feedback. Work in the Stern-Ginossar lab on m6A is supported by a European Research Council starting grant (grant no. StG-2014-638142) and by the Minerva foundation. The Shulman lab is supported by the ERC grant (grant no. StG-2015-677713), Human Frontiers of Science Program (grant no. CDA-00023/2016), Israel Science Foundation (ISF) grant no. 1090/18 and the Morris Kahn Institute for Human Immunology. N.S.-G is an incumbent of the Skirball New Scientist Chair. N.S.-G. and Z.S. are members of the European Molecular Biology Organization (EMBO) Young Investigator Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Editor recognition statement J. D. K. Wilson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shulman, Z., Stern-Ginossar, N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat Immunol 21, 501–512 (2020). https://doi.org/10.1038/s41590-020-0650-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-020-0650-4
This article is cited by
-
Regulation of inflammatory diseases via the control of mRNA decay
Inflammation and Regeneration (2024)
-
Exploring the prognostic potential of m6A methylation regulators in low-grade glioma: implications for tumor microenvironment modulation
European Journal of Medical Research (2024)
-
RNA methylation patterns, immune characteristics, and autophagy-related mechanisms mediated by N6-methyladenosine (m6A) regulatory factors in venous thromboembolism
BMC Genomics (2024)
-
Comprehensive analysis of m6A modification in immune infiltration, metabolism and drug resistance in hepatocellular carcinoma
Cancer Cell International (2024)
-
RNA N6-methyladenosine modifications in urological cancers: from mechanism to application
Nature Reviews Urology (2024)