Abstract
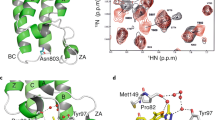
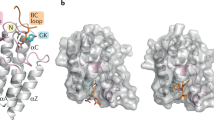
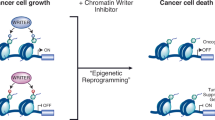
Chromatin regulatory proteins are increasingly recognized as potential new drug targets. Many of these proteins harbor one or more so called ‘reader domains’ that recognize covalent modifications of lysine and arginine residues, typically on histones, which mediate specific interactions within chromatin. Here we review recent progress in the discovery of drug-like small molecules that antagonize the function of methyl-lysine and methyl-arginine reader domains (Royal family, plant homeodomain (PHD) and WD40 domains) as well as the acyl-lysine-binding YEATS domain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Jeltsch, A., Broche, J. & Bashtrykov, P. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes 9, E566 (2018).
Zaware, N. & Zhou, M.-M. Bromodomain biology and drug discovery. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-019-0309-8 (2019).
Maurer-Stroh, S. et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 (2003).
Gayatri, S. & Bedford, M. T. Readers of histone methylarginine marks. Biochim. Biophys. Acta 1839, 702–710 (2014).
Collins, R. E. et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 15, 245–250 (2008).
Sanchez, R. & Zhou, M.-M. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36, 364–372 (2011).
Schapira, M., Tyers, M., Torrent, M. & Arrowsmith, C. H. WD40 repeat domain proteins: a novel target class? Nat. Rev. Drug Discov. 16, 773–786 (2017).
Trojer, P. et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915–928 (2007).
Nady, N. et al. Histone recognition by human malignant brain tumor domains. J. Mol. Biol. 423, 702–718 (2012).
Min, J. et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol. 14, 1229–1230 (2007).
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Meier, K. & Brehm, A. Chromatin regulation: how complex does it get? Epigenetics 9, 1485–1495 (2014).
Santiago, C., Nguyen, K. & Schapira, M. Druggability of methyl-lysine binding sites. J. Comput. Aided Mol. Des. 25, 1171–1178 (2011).
Herold, J. M. et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 54, 2504–2511 (2011).
James, L. I. et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 9, 184–191 (2013). First biologically active antagonist of a Kme reader domain, demonstrating proof of concept for this class of protein.
Lu, J. et al. L3MBTL1 regulates ALS/FTD-associated proteotoxicity and quality control. Nat. Neurosci. 22, 875–886 (2019).
Kaustov, L. et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 (2011).
Ren, C. et al. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem. Biol. 22, 161–168 (2015).
Simhadri, C. et al. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7). J. Med. Chem. 57, 2874–2883 (2014).
Stuckey, J. I. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat. Chem. Biol. 12, 180–187 (2016).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Lee, J., Thompson, J. R., Botuyan, M. V. & Mer, G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15, 109–111 (2008).
Roy, S. et al. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. J. Mol. Biol. 398, 489–496 (2010).
Bian, C. et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 (2011).
Arita, K. et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl Acad. Sci. USA 109, 12950–12955 (2012).
Tripsianes, K. et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420 (2011).
Liu, K. et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl Acad. Sci. USA 107, 18398–18403 (2010).
Sikorsky, T. et al. Recognition of asymmetrically dimethylated arginine by TDRD3. Nucleic Acids Res. 40, 11748–11755 (2012).
Perfetti, M. T. et al. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 10, 1072–1081 (2015).
Jurkowska, R. Z. et al. H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1. Nat. Commun. 8, 2057 (2017).
Mader, P. et al. Identification and characterization of the first fragment hits for SETDB1 Tudor domain. Bioorg. Med. Chem. 27, 3866–3878 (2019).
Senisterra, G. et al. Discovery of small-molecule antagonists of the H3K9me3 binding to UHRF1 tandem Tudor domain. SLAS Discov. 23, 930–940 (2018).
Houliston, R. S. et al. Conformational dynamics of the TTD-PHD histone reader module of the UHRF1 epigenetic regulator reveals multiple histone-binding states, allosteric regulation, and druggability. J. Biol. Chem. 292, 20947–20959 (2017).
Su, X. et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 28, 622–636 (2014).
Fagan, V. et al. A chemical probe for Tudor domain protein Spindlin1 to investigate chromatin functions. https://doi.org/10.26434/chemrxiv.7673129.v2 (2019). First-in-class chemical probe for a Tudor domain protein.
Wagner, T. et al. Identification of a small-molecule ligand of the epigenetic reader protein Spindlin1 via a versatile screening platform. Nucleic Acids Res. 44, e88 (2016).
Sweis, R. F. et al. Discovery and development of potent and selective inhibitors of histone methyltransferase g9a. ACS Med. Chem. Lett. 5, 205–209 (2014).
Bae, N. et al. Developing Spindlin1 small-molecule inhibitors by using protein microarrays. Nat. Chem. Biol. 13, 750–756 (2017).
Xiong, Y. et al. Discovery of a potent and selective fragment-like inhibitor of methyllysine reader protein spindlin 1 (SPIN1). J. Med. Chem. (2019).
Qin, S. & Min, J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 39, 536–547 (2014).
Böttcher, J. et al. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat. Chem. Biol. 15, 822–829 (2019). First-in-class PWWP inhibitor, with demonstrated impact on relevant biology.
Shen, C. et al. NSD3-Short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell 60, 847–859 (2015).
Klein, B. J. et al. Recognition of histone H3K14 acylation by MORF. Structure 25, 650–654.e2 (2017).
Amato, A., Lucas, X., Bortoluzzi, A., Wright, D. & Ciulli, A. Targeting ligandable pockets on plant homeodomain (PHD) zinc finger domains by a fragment-based approach. ACS Chem. Biol. 13, 915–921 (2018).
Miller, T. C. R. et al. Competitive binding of a benzimidazole to the histone-binding pocket of the Pygo PHD finger. ACS Chem. Biol. 9, 2864–2874 (2014).
Wagner, E. K., Nath, N., Flemming, R., Feltenberger, J. B. & Denu, J. M. Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry 51, 8293–8306 (2012).
Horton, J. R. et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17, 38–43 (2010).
Andrews, F. H. et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 12, 396–398 (2016).
Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 (2016).
Zhao, D. et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 26, 629–632 (2016).
Moustakim, M. et al. Discovery of an MLLT1/3 YEATS domain chemical probe. Angew. Chem. Int. Edn Engl. 57, 16302–16307 (2018).
Li, X. et al. Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking. Nat. Chem. Biol. 14, 1140–1149 (2018).
Schuetz, A. et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 25, 4245–4252 (2006).
Couture, J.-F., Collazo, E. & Trievel, R. C. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 13, 698–703 (2006).
Ruthenburg, A. J. et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13, 704–712 (2006).
Han, Z. et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell 22, 137–144 (2006).
Song, J.-J. & Kingston, R. E. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 283, 35258–35264 (2008).
Avdic, V. et al. Structural and biochemical insights into MLL1 core complex assembly. Structure 19, 101–108 (2011).
Patel, A., Vought, V. E., Dharmarajan, V. & Cosgrove, M. S. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 283, 32162–32175 (2008).
Dias, J. et al. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 28, 929–942 (2014).
Grebien, F. et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 11, 571–578 (2015). First small-molecule chemical probe to a chromatin WDR protein with demonstrated activity on target biology.
Aho, E. R. et al. Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity. Cell Rep. 26, 2916–2928.e13 (2019).
Karatas, H. et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 135, 669–682 (2013). First cell-permeant modulator of a chromatin-associated WDR domain with effects on relevant biology.
Karatas, H. et al. Discovery of a highly potent, cell-permeable macrocyclic peptidomimetic (MM-589) targeting the WD repeat domain 5 protein (WDR5)-mixed lineage leukemia (MLL) protein-protein interaction. J. Med. Chem. 60, 4818–4839 (2017).
Zhu, J. et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 (2015).
Xu, C. et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl Acad. Sci. USA 107, 19266–19271 (2010).
Lee, C.-H. et al. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol. Cell 70, 422–434.e6 (2018).
Justin, N. et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 (2016).
Jiao, L. & Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015).
He, Y. et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol. 13, 389–395 (2017). First (along with ref. 72) small-molecule allosteric modulation of a chromatin enzyme.
Qi, W. et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 13, 381–388 (2017). First (along with ref. 71) small-molecule allosteric modulation of a chromatin enzyme complex by targeting its Kme reader domain.
Suh, J. L. et al. Discovery of selective activators of PRC2 mutant EED-I363M. Sci. Rep. 9, 6524 (2019).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Arrowsmith, C.H., Schapira, M. Targeting non-bromodomain chromatin readers. Nat Struct Mol Biol 26, 863–869 (2019). https://doi.org/10.1038/s41594-019-0290-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0290-2
This article is cited by
-
Cytoskeleton remodeling induced by SMYD2 methyltransferase drives breast cancer metastasis
Cell Discovery (2024)
-
An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering
Nature Communications (2023)
-
Pharmacological perturbation of the phase-separating protein SMNDC1
Nature Communications (2023)
-
The PHD transcription factor Cti6 is involved in the fungal colonization and aflatoxin B1 biological synthesis of Aspergillus flavus
IMA Fungus (2021)
-
Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery
Scientific Reports (2021)