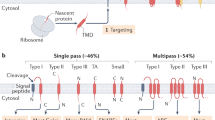
Abstract
Members of the Oxa1 superfamily perform membrane protein insertion in bacteria, the eukaryotic endoplasmic reticulum (ER), and endosymbiotic organelles. Here, we review recent structures of the three ER-resident insertases and discuss the extent to which structure and function are conserved with their bacterial counterpart YidC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Guna, A. & Hegde, R. S. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr. Biol. 28, R498–R511 (2018).
Chen, Y. & Dalbey, R. E. Oxa1 superfamily: new members found in the ER. Trends Biochem. Sci. 43, 151–153 (2018).
Hennon, S. W., Soman, R., Zhu, L. & Dalbey, R. E. YidC/Alb3/Oxa1 family of insertases. J. Biol. Chem. 290, 14866–14874 (2015).
Kumazaki, K. et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 (2014). This paper presents the first high-resolution structure of YidC and proposes a model for substrate insertion via the hydrophilic groove.
Kumazaki, K. et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4, 7299 (2014).
Anghel, S. A., McGilvray, P. T., Hegde, R. S. & Keenan, R. J. Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 21, 3708–3716 (2017). This paper identifies Get1, EMC3, and TMCO1 as ER homologues of YidC, thereby defining the Oxa1 superfamily.
McDowell, M. A. et al. Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell 80, 72–86.e7 (2020). This paper presents the cryo-EM structure of the human Get1–Get2–Get3 complex, identifying a membrane heterotetramer with two distinct hydrophilic grooves.
Pleiner, T. et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science 369, 433–436 (2020). This work presents the cryo-EM structure of the human EMC, identifying EMC3 as the site for insertion and the hydrophilic groove as functionally important.
Bai, L., You, Q., Feng, X., Kovach, A. & Li, H. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 584, 475–478 (2020). This paper presents the cryo-EM structure of yeast EMC, identifying the hydrophilic groove as functionally important for substrate insertion.
O'Donnell, J. P. et al. The architecture of EMC reveals a path for membrane protein insertion. Elife 9, e57887 (2020). This study presents the cryo-EM structure of human EMC and a crystal structure of EMC2, identifying it as an interaction site for substrate TMDs.
Miller-Vedam, L. E. et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. Elife 9, e62611 (2020). This paper presents the structures of the yeast and human EMC, identifying two lipid-accessible membrane cavities involved in terminal helix insertion and polytopic membrane protein biogenesis.
McGilvray, P. T. et al. An ER translocon for multi-pass membrane protein biogenesis. Elife 9, e56889 (2020). This paper presents the structure of the TMCO1 translocon and shows that it is important for membrane insertion of substrates with four or more TMDs.
van Bloois, E. et al. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J. Biol. Chem. 280, 12996–13003 (2005).
Jiang, F. et al. Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J. Biol. Chem. 277, 19281–19288 (2002).
Laan, M. V. D., Nouwen, N. P. & Driessen, A. J. M. YidC — an evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr. Opin. Microbiol. 8, 182–187 (2005).
Walter, B., Hristou, A., Nowaczyk, Marc, M. & Schünemann, D. In vitro reconstitution of co-translational D1 insertion reveals a role of the cpSec–Alb3 translocase and Vipp1 in photosystem II biogenesis. Biochem. J. 468, 315–324 (2015).
Samuelson, J. C. et al. YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 (2000).
Borgese, N., Coy-Vergara, J., Colombo, S. F. & Schwappach, B. The ways of tails: the GET pathway and more. Protein J. 38, 289–305 (2019).
Guna, A., Volkmar, N., Christianson, J. C. & Hegde, R. S. The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473 (2018).
Chitwood, P. J., Juszkiewicz, S., Guna, A., Shao, S. & Hegde, R. S. EMC is required to initiate accurate membrane protein topogenesis. Cell 175, 1507–1519 (2018).
Shurtleff, M. J. et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife 7, e37018 (2018).
Wang, F., Chan, C., Weir, N. R. & Denic, V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 512, 441–444 (2014).
Schuldiner, M. et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 (2008).
Xin, B. et al. Homozygous frameshift mutation in TMCO1 causes a syndrome with craniofacial dysmorphism, skeletal anomalies, and mental retardation. Proc. Natl Acad. Sci. USA 107, 258–263 (2010).
Xin, Y. et al. Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion. FASEB J. 32, 2411–2421 (2018).
Tanaka, Y. et al. 2.8-Å crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop. Biochem. Biophys. Res. Commun. 505, 141–145 (2018).
Lemaire, C., Guibet-Grandmougin, F., Angles, D., Dujardin, G. & Bonnefoy, N. A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J. Biol. Chem. 279, 47464–47472 (2004).
Chen, Y. et al. YidC insertase of Escherichia coli: water accessibility and membrane shaping. Structure 25, 1403–1414 (2017).
Klenner, C. & Kuhn, A. Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts. J. Biol. Chem. 287, 3769–3776 (2012).
Yu, Z., Koningstein, G., Pop, A. & Luirink, J. The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J. Biol. Chem. 283, 34635–34642 (2008).
Wickles, S. et al. A structural model of the active ribosome-bound membrane protein insertase YidC. Elife 3, e03035 (2014).
Hariharan, B. et al. Polarity/charge as a determinant of translocase requirements for membrane protein insertion. Biochim. Biophys. Acta 1863, 183502 (2021).
Rodriguez, F. et al. Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl Acad. Sci. USA 110, E1092–E1101 (2013).
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013).
Wu, X. et al. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368, eaaz2449 (2020).
Gafvelin, G., Sakaguchi, M., Andersson, H. & von Heijne, G. Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem. 272, 6119–6127 (1997).
Baker, J. A., Wong, W.-C., Eisenhaber, B., Warwicker, J. & Eisenhaber, F. Charged residues next to transmembrane regions revisited: “positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol. 15, 66 (2017).
Kedrov, A. et al. Structural dynamics of the YidC:ribosome complex during membrane protein biogenesis. Cell Rep. 17, 2943–2954 (2016).
Dalbey, R. E. & Kuhn, A. How YidC inserts and folds proteins across a membrane. Nat. Struct. Mol. Biol. 21, 435–436 (2014).
He, H., Kuhn, A. & Dalbey, R. E. Tracking the stepwise movement of a membrane-inserting protein in vivo. J. Mol. Biol. 432, 484–496 (2020).
Wang, F., Whynot, A., Tung, M. & Denic, V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol. Cell 43, 738–750 (2011).
Yamamoto, Y. & Sakisaka, T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 48, 387–397 (2012).
Zalisko, B. E., Chan, C., Denic, V., Rock, R. S. & Keenan, R. J. Tail-anchored protein insertion by a single Get1/2 heterodimer. Cell Rep. 20, 2287–2293 (2017).
Boy, D. & Koch, H.-G. Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815 (2009).
Kohler, R. et al. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell 34, 344–353 (2009).
Spann, D., Pross, E., Chen, Y., Dalbey, R. E. & Kuhn, A. Each protomer of a dimeric YidC functions as a single membrane insertase. Sci. Rep. 8, 589 (2018).
Volkmar, N. et al. The ER membrane protein complex promotes biogenesis of sterol-related enzymes maintaining cholesterol homeostasis. J. Cell Sci. 132, jcs223453 (2019).
Sachelaru, I. et al. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 288, 16295–16307 (2013).
Botte, M. et al. A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci. Rep. 6, 38399 (2016).
Urbanus, M. L. et al. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 (2001).
Petriman, N.-A. et al. The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci. Rep. 8, 578 (2018).
Klostermann, E., Droste Gen Helling, I., Carde, J.-P. & Schünemann, D. The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem. J. 368, 777–781 (2002).
Stefer, S. et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3–receptor complex. Science 333, 758–762 (2011).
Mariappan, M. et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477, 61–66 (2011).
Bozkurt, G. et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl Acad. Sci. USA 106, 21131–21136 (2009).
Hu, J., Li, J., Qian, X., Denic, V. & Sha, B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One 4, e8061 (2009).
Mateja, A. et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361–366 (2009).
Suloway, C. J. M., Chartron, J. W., Zaslaver, M. A. & Clemons, W. M. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl Acad. Sci. USA 106, 14849–14854 (2009).
Mateja, A. et al. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347, 1152–1155 (2015).
Kubota, K., Yamagata, A., Sato, Y., Goto-Ito, S. & Fukai, S. Get1 stabilizes an open dimer conformation of Get3 ATPase by binding two distinct interfaces. J. Mol. Biol. 422, 366–375 (2012).
Rome, M. E., Chio, U. S., Rao, M., Gristick, H. & Shan, S.-O. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc. Natl Acad. Sci. USA 111, 4929–4935 (2014).
Denic, V., Dotsch, V. & Sinning, I. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb. Perspect. Biol. 5, a013334 (2013).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Hennon, S. W. & Dalbey, R. E. Cross-linking-based flexibility and proximity relationships between the TM segments of the Escherichia coli YidC. Biochemistry 53, 3278–3286 (2014).
Borowska, M. T., Dominik, P. K., Anghel, S. A., Kossiakoff, A. A. & Keenan, R. J. A YidC-like protein in the archaeal plasma membrane. Structure 23, 1715–1724 (2015).
Kedrov, A. et al. Elucidating the native architecture of the YidC: ribosome complex. J. Mol. Biol. 425, 4112–4124 (2013).
Seitl, I., Wickles, S., Beckmann, R., Kuhn, A. & Kiefer, D. The C-terminal regions of YidC from Rhodopirellula baltica and Oceanicaulis alexandrii bind to ribosomes and partially substitute for SRP receptor function in Escherichia coli. Mol. Microbiol. 91, 408–421 (2014).
Welte, T. et al. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479 (2012).
Falk, S., Ravaud, S., Koch, J. & Sinning, I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962 (2010).
Jia, L. et al. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22, 6438–6447 (2003).
Szyrach, G., Ott, M., Bonnefoy, N., Neupert, W. & Herrmann, J. M. Ribosome binding to the Oxa1 complex facilitates co‐translational protein insertion in mitochondria. EMBO J. 22, 6448–6457 (2003).
Dong, R., Pan, S., Peng, Z., Zhang, Y. & Yang, J. mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Res. 46, W380–W386 (2018).
Acknowledgements
This work was supported by an EMBO Long Term Fellowship (ALTF 1230-2013) to M.A.M. and by the DFG through the Leibniz Programme (SI 586/6-1) and TRR83 (TP22) to I.S.
Author information
Authors and Affiliations
Contributions
M.A.M., M.H. and I.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
McDowell, M.A., Heimes, M. & Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat Struct Mol Biol 28, 234–239 (2021). https://doi.org/10.1038/s41594-021-00567-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00567-9
This article is cited by
-
EMC rectifies the topology of multipass membrane proteins
Nature Structural & Molecular Biology (2024)
-
The GET insertase exhibits conformational plasticity and induces membrane thinning
Nature Communications (2023)
-
Der mysteriöse SND-Weg beim Membranproteintransport
BIOspektrum (2023)
-
The mechanisms of integral membrane protein biogenesis
Nature Reviews Molecular Cell Biology (2022)
-
Role of a bacterial glycolipid in Sec-independent membrane protein insertion
Scientific Reports (2022)