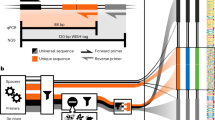
Abstract
Commensal bacteria from the human intestinal microbiota play important roles in health and disease. Research into the mechanisms by which these bacteria exert their effects is hampered by the complexity of the microbiota, the strict growth requirements of the individual species and a lack of genetic tools and resources. The assembly of ordered transposon insertion libraries, in which nearly all nonessential genes have been disrupted and the strains stored as independent monocultures, would be a transformative resource for research into many microbiota members. However, assembly of these libraries must be fast and inexpensive in order to empower investigation of the large number of species that typically compose gut communities. The methods used to generate ordered libraries must also be adapted to the anaerobic growth requirements of most intestinal bacteria. We have developed a protocol to assemble ordered libraries of transposon insertion mutants that is fast, cheap and effective for even strict anaerobes. The protocol differs from currently available methods by making use of cell sorting to order the library and barcoded transposons to facilitate the localization of ordered mutations in the library. By tracking transposon insertions using barcode sequencing, our approach increases the accuracy and reduces the time and effort required to locate mutants in the library. Ordered libraries can be sorted and characterized over the course of 2 weeks using this approach. We expect this protocol will lower the barrier to generating comprehensive, ordered mutant libraries for many species in the human microbiota, allowing for new investigations into genotype–phenotype relationships within this important microbial ecosystem.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequencing data used in this study are publicly available on NCBI as part of BioProject PRJNA573294. The sequence reads from the Bar-seq experiment on pools of the ordered library are available as part of BioSample SAMN12807646. The sequence reads from the RB-TnSeq experiment on the ordered library are available as part of BioSample SAMN12809978. Plate reader output files, microscopy images and extracted growth parameters that support the findings of this study (Supplementary Figs. 2–4) are available from the corresponding author upon request.
Code availability
The code required for locating insertion mutants in ordered libraries is available on BitBucket (https://bitbucket.org/kchuanglab/resolve_barcode_position/src). These scripts rely on previously published code available on BitBucket (https://bitbucket.org/berkeleylab/feba/src). The code in this protocol has been peer reviewed.
References
Gentile, C. L. & Weir, T. L. The gut microbiota at the intersection of diet and human health. Science 362, 776–780 (2018).
Tramontano, M. et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522 (2018).
Liberati, N. T. et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl Acad. Sci. 103, 2833–2838 (2006).
De Berardinis, V. et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4, 174 (2008).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009).
Porwollik, S. et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS ONE 9, e99820 (2014).
Vandewalle, K. et al. Characterization of genome-wide ordered sequence-tagged Mycobacterium mutant libraries by Cartesian Pooling-Coordinate Sequencing. Nat. Commun. 6, 7106 (2015).
Baym, M., Shaket, L., Anzai, I. A., Adesina, O. & Barstow, B. Rapid construction of a whole-genome transposon insertion collection for Shewanella oneidensis by Knockout Sudoku. Nat. Commun. 7, 13270 (2016).
Koo, B.-M. et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4, 291–305.e297 (2017).
Dale, J. L. et al. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3, e00062–00018 (2018).
Baba, T. & Mori, H. The construction of systematic in-frame, single-gene knockout mutant collection in Escherichia coli K-12. Methods Mol. Biol. 416, 171–181 (2008).
Lampe, D. J., Akerley, B. J., Rubin, E. J., Mekalanos, J. J. & Robertson, H. M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl Acad. Sci. 96, 11428–11433 (1999).
Goryshin, I. Y. & Reznikoff, W. S. Tn5 in vitro transposition. J. Biol. Chem. 273, 7367–7374 (1998).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648 (2017).
Goodman, A. L., Wu, M. & Gordon, J. I. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 6, 1969 (2011).
Anzai, I. A., Shaket, L., Adesina, O., Baym, M. & Barstow, B. Rapid curation of gene disruption collections using Knockout Sudoku. Nat. Protoc. 12, 2110 (2017).
Erlich, Y. et al. DNA Sudoku—harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 19, 1243–1253 (2009).
Wetmore, K. M. et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6, e00306–e00315 (2015).
Price, M. N. et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503 (2018).
Hughes, S. R. et al. High-throughput screening of cellulase F mutants from multiplexed plasmid sets using an automated plate assay on a functional proteomic robotic workcell. Proteome Sci. 4, 10 (2006).
Liu, H. et al. Functional genetics of human gut commensal Bacteroides thetaiotaomicron reveals metabolic requirements for growth across environments. Cell Rep 34, 108789 (2021).
Shatalin, K., Shatalina, E., Mironov, A. & Nudler, E. H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 (2011).
Holt, J.G., Kreig, N. R., Sneath, P. H. A., Staley J. T. & Williams S. T. Bergey’s Manual of Determinative Bacteriology 9th edn (Williams & Willkins, 1994).
Acknowledgements
The authors thank the Huang lab for helpful discussions. This work was supported in part by the Allen Discovery Center at Stanford University on Systems Modeling of Infection (to K.C.H.) and NIH grants RM1 GM135102 (to A.M.D. and K.C.H.) and R01 AI147023 (to K.C.H.). K.C.H. is a Chan Zuckerberg Biohub Investigator. The authors also acknowledge the hospitality of the Aspen Center for Physics, which is supported by NSF grant PHY-1607611. The following reagents were obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project: Lactobacillus reuteri, strain CF48-3A, HM-102; Clostridium innocuum, strain 6_1_30, HM-173; Clostridium symbiosum, strain WAL-14163, HM-309; Bacteroides finegoldii, strain CL09T03C10, HM-727; Parabacteroides johnsonii, strain CL02T12C29, HM-731.
Author information
Authors and Affiliations
Contributions
A.L.S., A.M.D. and K.C.H. conceived the study. A.L.S. designed experiments and collected data. A.L.S. and R.C. analyzed the data. A.L.S., R.C., A.M.D. and K.C.H. interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Buz Barstow and Matthew Waldor for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Price, M. N. et al. Nature 557, 503 (2018): https://doi.org/10.1038/s41586-018-0124-0
Liu, H. et al. Cell Rep. 34, 108789 (2021): https://doi.org/10.1016/j.celrep.2021.108789
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Supplementary Results and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Shiver, A.L., Culver, R., Deutschbauer, A.M. et al. Rapid ordering of barcoded transposon insertion libraries of anaerobic bacteria. Nat Protoc 16, 3049–3071 (2021). https://doi.org/10.1038/s41596-021-00531-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00531-3
This article is cited by
-
High-throughput anaerobic screening for identifying compounds acting against gut bacteria in monocultures or communities
Nature Protocols (2024)
-
Double emulsions as a high-throughput enrichment and isolation platform for slower-growing microbes
ISME Communications (2023)
-
Functional annotation and importance of marine bacterial transporters of plankton exometabolites
ISME Communications (2023)
-
Profiling the human intestinal environment under physiological conditions
Nature (2023)
-
Construction and characterization of a genome-scale ordered mutant collection of Bacteroides thetaiotaomicron
BMC Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.