Abstract
Lignin is a phenylpropanoid polymer produced in the secondary cell walls of vascular plants. Although most eudicot and gymnosperm species generate lignins solely via polymerization of p-hydroxycinnamyl alcohols (monolignols), grasses additionally use a flavone, tricin, as a natural lignin monomer to generate tricin-incorporated lignin polymers in cell walls. We previously found that disruption of a rice 5-HYDROXYCONIFERALDEHYDE O-METHYLTRANSFERASE (OsCAldOMT1) reduced extractable tricin-type metabolites in rice vegetative tissues. This same enzyme has also been implicated in the biosynthesis of sinapyl alcohol, a monolignol that constitutes syringyl lignin polymer units. Here, we further demonstrate through in-depth cell wall structural analyses that OsCAldOMT1-deficient rice plants produce altered lignins largely depleted in both syringyl and tricin units. We also show that recombinant OsCAldOMT1 displayed comparable substrate specificities towards both 5-hydroxyconiferaldehyde and selgin intermediates in the monolignol and tricin biosynthetic pathways, respectively. These data establish OsCAldOMT1 as a bifunctional O-methyltransferase predominantly involved in the two parallel metabolic pathways both dedicated to the biosynthesis of tricin-lignins in rice cell walls. Given that cell wall digestibility was greatly enhanced in the OsCAldOMT1-deficient rice plants, genetic manipulation of CAldOMTs conserved in grasses may serve as a potent strategy to improve biorefinery applications of grass biomass.
Similar content being viewed by others
Introduction
Grasses, including cereals classified in the major monocot family Poaceae, show great potential as a source of lignocellulosic biomass, which is primarily composed of secondary cell walls produced in vascular tissues. A large amount of lignocellulosic biomass is produced worldwide annually as agricultural residues from grass grain crops including maize, wheat, rice, barley and sorghum. In addition, the so-called grass biomass crops, such as Miscanthus, Erianthus, switchgrass, and bamboos, are attracting attention as new sources of biomass for the production of biofuels and biochemicals, especially because of their superior lignocellulosic productivity and processability when compared with those of typical softwood and hardwood species, currently used in the timber and pulp industries1,2. To further improve our capacity to manipulate grass biomass by molecular breeding approaches, it is becoming increasingly important to deepen our understanding of the formation, structure, and properties of secondary cell walls in grasses2,3,4.
Lignin is a complex phenylpropanoid polymer derived from oxidative coupling of p-hydroxycinnamyl alcohols, namely monolignols, and related compounds, and typically accounts for 15%–30% of lignocellulosic material. By encrusting cell wall polysaccharides, i.e., cellulose and hemicelluloses, lignin confers enhanced mechanical strength, imperviousness, and resistance to pathogens5,6,7. Although the polymer is vital for all vascular plants to maintain their essential functions of water transport and structural support, lignin has long been considered a major obstacle for polysaccharide-oriented biomass utilization, including chemical pulping and fermentable sugar production. Accordingly, lignin bioengineering studies have traditionally targeted plants with reduced lignin content and/or altered lignin chemical structure to mitigate such lignin recalcitrance in polysaccharide utilization processes5,6,7,8. More recently, as lignin has been increasingly viewed as a viable source for biomass-derived aromatic fuels and commodities, alternative bioengineering approaches that manipulate lignin content/structure to improve lignin-oriented biomass utilization strategies are also becoming an important research subject2,9,10.
There is accumulating evidence that the structure and biosynthesis of lignins in grasses are distinctively different from those in gymnosperms (softwoods) and eudicots (including hardwoods)11. As a prime example, many grasses appear to use tricin, a 3′,5′-dimethoxyflavone, for cell wall lignification, which contrasts gymnosperm and eudicot species that typically use the canonical monolignols as sole lignin monomers12,13,14. Tricin, as a natural lignin monomer generated outside the monolignol biosynthetic pathway (Fig. 1), undergoes dehydrogenative co-polymerization with monolignols exclusively via 4′–O–β-type radical coupling upon cell wall lignification in grasses, as in the way lignification takes place solely with monolignols in typical gymnosperms and eudicots13. Such tricin-incorporated lignins, or tricin-lignins, have been shown to exist commonly in grasses and are also found in some non-grass monocots such as curaua, coconut, and orchids, as well as in the dicot alfalfa, albeit at very low levels14. Although it has long been recognized that monolignol-derived lignins and flavonoids generally localize and function differently in plants5,15,16, the discovery of tricin-lignins indicates a tight relationship between the two major classes of plant metabolite derived from the phenylpropanoid pathway; however, it remains largely unknown how such tricin-lignins are biosynthesized and function in grass cell walls. Moreover, how the existence of tricin-lignins affects the usability of grass biomass remains poorly understood.
Proposed monolignol and tricin biosynthetic pathways in rice. HCT, p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase; C3′H, p-coumaroyl ester 3-hydroxylase; CSE, caffeoyl shikimate esterase; 4CL: 4-coumaroyl-CoA ligase; cinnamoyl-CoA reductase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAld5H, coniferaldehyde 5-hydroxylase; CAldOMT, 5-hydroxyconiferaldehyde O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; PMT, p-coumaroyl-CoA:monolignol transferase; CHS, chalcone synthase; CHI, chalcone isomerase; FNSII, flavone synthase II; A3′H/C5′H, apigenin 3′-hydroxylase/chrysoeriol 5′-hydroxylase; OMT, O-methyltransferase; LAC, laccase; PRX, peroxidase.
Monolignols and tricin are both major downstream metabolites in the phenylpropanoid biosynthetic pathway in grasses. In general, after branching off from p-coumaroyl-CoA (pCA-CoA), a common precursor in the monolignol and flavonoid biosynthetic pathways, monolignols are synthesized through successive aromatic hydroxylations and O-methylations, along with side-chain reductions and occasionally with further side-chain acylations. In grasses, the major monolignol-type lignin monomers are p-coumaryl, coniferyl, and sinapyl alcohols, and their γ-p-coumaroylated derivatives, which together make up the canonical p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units in lignin polymers, respectively, in cell walls (Fig. 1)5,17. On the other hand, the biosynthesis of tricin may involve the formation of the flavone backbone via the condensation of pCA-CoA with malonyl-CoA followed by chalcone isomerization and desaturation, leading to apigenin which is then converted into tricin through successive aromatic hydroxylations and O-methylations on the B-ring (Fig. 1). We recently identified some flavone biosynthetic enzymes responsible for the formation of tricin-lignins in rice cell walls by thorough characterization of rice mutant lines; rice mutants deficient in a rice FLAVONE SYNTHASE II (OsFNSII or CYP93G1)18 and a bifunctional APIGENIN 3′-HYDROXYLASE/CHRYSOERIOL 5′-HYDROXYLASE (OsA3′H/C5′H or CYP75B4)19 (Fig. 1) produced altered lignins completely devoid of tricin units and partially incorporating naringenin and apigenin, respectively, as a tricin surrogate, demonstrating predominant roles of these enzyme genes in tricin-lignin formation in rice cell walls. Both FNSII and A3′H/C5′H sequences appear to be highly conserved among grasses and likely function in the synthesis of tricin-lignins commonly present in grass cell walls18,19.
5-HYDROXYCONIFERALDEHYDE O-METHYLTRANSFERASE (CAldOMT or CAFFEIC ACID O-METHYLTRANSFERASE, COMT) is a key enzyme involved in the biosynthesis of S lignins in angiosperm species. In general, recombinant CAldOMTs accept many types of phenylpropanoid substrates but usually show notably higher catalytic efficiencies toward 5-hydroxyconiferaldehyde, a possible in planta substrate in the S lignin biosynthetic pathway that has diverged from the G lignin biosynthetic pathway20,21,22,23,24 (Fig. 1). In agreement with the proposed function of CAldOMT in S lignin biosynthesis, downregulation of CAldOMT typically results in reduced S lignin content often accompanied by the appearance of atypical 5-hydroxyguaiacyl (5H) lignin units via incorporation of the non-canonical 5-hydroxyconiferyl alcohol monomer upon lignification23,24,25,26,27,28,29 (Fig. 1). In some grasses, such as maize and sorghum, mutations in CAldOMT genes are associated with a brown midrib (bm) phenotype that often exhibits reduced lignin content and enhanced forage digestibility30,31. Similarly to such bm mutants, transgenic plants downregulated for CAldOMT typically display enhanced forage digestibility and/or biomass saccharification efficiency23,26,31,32,33,34,35,36,37,38. Hence, CAldOMT has been considered one of the promising targets in lignin bioengineering studies.
Previously, we demonstrated that a rice CAldOMT, OsCAldOMT1 (also known as OsCOMT1 or ROMT9), is requisite for S lignin biosynthesis in rice, a grass model species and a commercially important crop; an RNA-interference (RNAi)-derived OsCAldOMT1-knockdown rice (OsCAldOMT1-RNAi) was significantly depleted in S lignin units with increments in G and 5H lignin units23. Interestingly, however, in a separate work, we also found that a T-DNA insertional mutant rice deficient in the same gene (OsCAldOMT1-TDNA) was remarkably reduced in some extractable flavone metabolites including tricin and its O-conjugates in the vegetative tissues39. These observations collectively suggest that OsCAldOMT1 may have a dual role along both the monolignol and tricin biosynthetic pathways, which together direct the formation of tricin-lignins in rice cell walls; however, the precise involvement of OsCAldOMT1 in tricin-lignin biosynthesis in rice has not been completely addressed.
In this study, we provide evidence for the bifunctional role of OsCAldOMT1 in the formation of tricin-lignins in rice through in-depth cell wall structural analysis using histochemical, chemical, and two-dimensional (2D) NMR methods on the OsCAldOMT1-deficient rice lines. We also performed comparative kinetic assays of a recombinant OsCAldOMT1 using 5-hydroxyconiferaldehyde and selgin, potential in planta substrates of OsCAldOMT1. Our data firmly establish the bifunctional role of OsCAldOMT1 in generating both S and tricin lignin polymer units in rice cell walls. Taken together with the corroborative cell wall NMR data recently reported for CAldOMT-deficient maize36 and sorghum40, both of which showed reduced tricin-lignin contents in their cell walls, the bifunctional role of CAldOMT in tricin-lignin biosynthesis may be well conserved in the grass family.
Results
Sequence and expression analysis of OsCAldOMT1
We first performed a phylogenetic analysis to examine the phylogenetic relationship between OsCAldOMT1 and other CAldOMT proteins implicated in lignification. As shown in Supplementary Fig. S1, OsCAldOMT1 was clustered with other previously characterized grass CAldOMTs, including those in Brachypodium distachyon35, maize36, barley38, switchgrass33, wheat41, sugarcane34 and sorghum40 with high sequence identities (79%–83% identity) and separated from dicot CAldOMTs (47%–61% identity) in Arabidopsis24,42,43,44, poplar species20,27, Medicago sativa26, and Nicotiana tabacum45, all of which have been established as bona fide CAldOMTs functioning in S lignin biosynthesis (Supplementary Fig. S1).
Next, we examined the spatio-temporal expression pattern of OsCAldOMT1 in wild-type rice (cv. Nipponbare) along with the other known/putative monolignol and tricin biosynthetic genes using the RiceXPro gene expression database46. OsCAldOMT1 is concurrently expressed with monolignol biosynthetic genes, such as p-COUMAROYL ESTER 3-HYDROXYLASE (OsC3′H1)47, CONIFERALDEHYDE 5-HYDROXYLASE (OsCAld5H1)48,49, CINNAMYL ALCOHOL DEHYDROGENASE (OsCAD2)50 and p-COUMAROYL-COA:MONOLIGNOL TRANSFERASE (OsPMT1)51 (Fig. 1), with high expression levels in tissues in which lignification typically occurs (Supplementary Fig. S2). In addition, OsCAldOMT1 shares a similar expression pattern with the known/putative tricin biosynthetic genes, including CHALCONE SYNTHASE (OsCHS1)52, CHALCONE ISOMERASE (OsCHI)52,53, FLAVONE SYNTHASE II (OsFNSII)18,54 and APIGENIN 3′-HYDROXYLASE/CHRYSOERIOL 5′-HYDROXYLASE (OsA3′H/C5′H)39 (Supplementary Fig. S2); among which OsFNSII18 and OsA3′H/C5′H19 have been shown to be indispensable for the formation of tricin-lignins in rice cell walls. These data are in line with our hypothesis that OsCAldOMT1 is involved in both the monolignol and tricin biosynthetic pathways in rice.
Enzyme assay of recombinant OsCAldOMT1
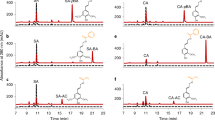
In our previous biochemical study, recombinant OsCAldOMT1 was found to catalyze O-methylation of several possible intermediates in the monolignol biosynthetic pathway, such as caffeic acid, caffeoyl-CoA, 5-hydroxyferuloyl-CoA, caffealdehyde, 5-hydroxyconiferaldehyde, caffeyl alcohol, and 5-hydroxyconiferyl alcohol, among which 5-hydroxyconiferaldehyde, a key intermediate in S lignin biosynthesis (Fig. 1), was the most preferential substrate23. Here, we extend the enzyme assay on selgin, which is a logical substrate of CAldOMT in the tricin biosynthetic pathway (Fig. 1). Recombinant OsCAldOMT123 produced tricin when incubated with selgin, demonstrating its capability to 5′-O-methylate selgin in vitro (Fig. 2a). Enzyme substrate specificity was then examined comparatively for 5-hydroxyconiferaldehyde and selgin. As a consequence, we found that the recombinant OsCAldOMT1 displayed comparable catalytic efficiencies (kcat/Km > 2.3 µM−1s−1) towards both 5-hydroxyconiferaldehyde and selgin (Fig. 2b). Overall, our data further corroborate the dual functions of OsCAldOMT1 in tricin-lignin biosynthesis in rice.
Kinetic assays for recombinant OsCAldOMT1. (a) HPLC chromatograms of reaction mixtures obtained for reactions of recombinant OsCAldOMT1 with 5-hydroxyconiferaldehyde (upper) and selgin (lower) substrates (detection by UV absorbance at 254 nm). (b) Kinetic parameters obtained for the reactions of recombinant OsCAldOMT1 with 5-hydroxyconiferaldehyde and selgin substrates. Values are means ± standard errors (n = 3).
Phenotypes of OsCAldOMT1-deficient rice
To further investigate the in planta role of OsCAldOMT1, we re-examined our previously generated OsCAldOMT1-deficient rice lines, namely, the OsCAldOMT1-RNAi knockdown (cv. Nipponbare)23 and OsCAldOMT1-TDNA knockout (cv. Hwayoung)39 lines. We previously determined that OsCAldOMT1 transcript levels were largely reduced down to ~1% of the wild-type levels in major vegetative tissues of OsCAldOMT1-RNAi23, whereas we expected a complete loss of functional OsCAldOMT1 activity in OsCAldOMT1-TDNA, which harbors a T-DNA insertion in the first exon of the OsCAldOMT1 locus39. Both OsCAldOMT1-RNAi and OsCAldOMT1-TDNA plants were grown side-by-side with their wild-type controls for phenotypic characterization (Supplementary Fig. S3; Table 1). Under the present growth conditions, both OsCAldOMT1-deficient lines did not show significant differences in their growth parameters when compared with those of the wild-type controls, except that slight reductions in plant height were observed for OsCAldOMT1-TDNA (Supplementary Fig. S3; Table 1). The notable growth differences between OsCAldOMT1-RNAi and OsCAldOMT1-TDNA and between their corresponding wild-type plants, i.e., between WT1 and WT2 (Table 1), were probably because of their different genetic backgrounds.
Histochemical analysis of OsCAldOMT1-deficient rice cell walls
Transverse sections from developing culms of OsCAldOMT1-RNAi and OsCAldOMT1-TDNA plants at heading stage were subject to cell wall histochemical analysis using phloroglucinol-HCl and vanillin-HCl reagents, which stain monolignol-derived lignins and flavonoids, respectively18. As visualized by phloroglucinol-HCl, the cell wall anatomy and the distribution of lignified tissues in OsCAldOMT1-RNAi and OsCAldOMT1-TDNA culms were overall similar to those observed in the wild-type controls, although we observed slightly reduced lignin signals in the cortical sclerenchyma fibers in OsCAldOMT1-RNAi and OsCAldOMT1-TDNA culms compared to in the wild-type control culms (Fig. 3). In contrast, whereas the lignified cortical sclerenchyma fiber and vascular bundle cell walls of wild-type culms displayed intense and positive yellowish colorations upon vanillin-HCl flavonoid staining, the flavonoid signal was clearly depleted in both OsCAldOMT1-RNAi and OsCAldOMT1-TDNA culm cell walls, suggesting that cell-wall-bound flavonoid, presumably lignin-bound tricin, was substantially reduced by the disruption of OsCAldOMT1 (Fig. 3).
Histochemical analysis of rice culm tissues of OsCAldOMT1-RNAi, OsCAldOMT1-TDNA and their wild-type controls (WT1 and WT2). Transverse sections of culm tissues were subjected to phloroglucinol-HCl and vanillin-HCl stains for visualization of monolignol-derived lignins and cell-wall-bound flavonoids, respectively.
Lignocellulosic compositional analysis of OsCAldOMT1-deficient rice cell walls
The impacts of OsCAldOMT1-deficiency on the lignocellulosic composition in rice cell walls were further examined by a series of wet-chemical analyses. The Klason lignin analysis on the rice culm cell wall residue (CWR) samples revealed that lignin content in the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls were reduced by 21% (from 142.4 to 112.6 mg/g CWR) and 30% (from 153.5 to 107.3 mg/g CWR) when compared with those of the wild-type controls, respectively (Table 2). In contrast, neutral-sugars analysis suggested that the amounts of arabinose and xylose released from hemicellulosic glycans were significantly increased in both OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls, whereas the amount of glucose released from crystalline cellulose remained constant; we also observed additional increments in amorphous glucans and galactans in the OsCAldOMT1-TDNA cell walls (Table 2). These data suggested that the lignin reductions in the OsCAldOMT1-deficient rice cell walls were at least partially compensated by relative increases in the levels of arabinoxylans.
We also quantified cell-wall-bound p-coumarate (pCA) and ferulate (FA) as the corresponding free acids released via mild alkaline hydrolysis. Both the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls were substantially depleted in pCA and conversely augmented in FA; approximately 56% (from 12.4 to 5.5 µmol/g CWR) and 59% (from 10.9 to 4.4 µmol/g CWR) reductions in pCA levels, and 62% (from 4.1 to 6.6 µmol/g CWR) and 39% (from 3.7 to 5.2 µmol/g CWR) increments in FA levels were observed for the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls, respectively (Table 2). Given that, in typical grass cell walls, the majority of pCA is bound to lignins whereas (releasable) FA is mainly associated with hemicelluloses, in particular, arabinoxylans (Ralph, 2010), these changes in pCA and FA levels in the OsCAldOMT1-deficient cell walls can be attributed to relatively reduced levels of lignins and increases in arabinoxylans, respectively (Table 2).
Lignin compositional analysis of OsCAldOMT1-deficient rice cell walls
We previously examined lignin composition in the OsCAldOMT1-RNAi23 using analytical thioacidolysis, which specifically cleaves β–O–4 linkages in lignin polymers and releases quantifiable monomeric compounds reflecting the polymer unit composition (Supplementary Fig. S4)55. Here, lignin composition in the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls was further evaluated by thioacidolysis, as well as derivatization followed by reductive cleavage (DFRC) methods. As DFRC also cleaves β–O–4 linkages in lignin polymers, but, unlike thioacidolysis, retains the γ-acyl groups and releases quantifiable γ-p-coumaroylated monomeric compounds along with non-acylated (γ-free) monomeric compounds, it provides useful information about the γ-acylation status of lignin polymers in grasses (Supplementary Fig. S4)56.
Both thioacidolysis and DFRC suggested that S/G ratios were drastically reduced in the OsCAldOMT1-deficient rice cell walls when compared with the wild-type controls: thioacidolysis-derived S/G ratios (Sthio/Gthio) were reduced by 85% (from 1.0 to 0.1) and 81% (from 1.1 to 0.2) and DFRC-derived S/G ratios (SDFRC-total/GDFRC-total; SDFRC-total = SDFRC-OH + SDFRC-pCA, GDFRC-total = GDFRC-OH + GDFRC-pCA) by 68% (from 0.7 to 0.2) and 52% (from 0.5 to 0.2) in the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls, respectively (Table 2; Supplementary Fig. S4), affirming that CAldOMT1 plays a key role in the biosynthesis of S lignins in rice23. DFRC further determined that both the non-acylated S (SDFRC-OH) and γ-p-coumaroylated S (SDFRC-pCA) monomeric compounds were proportionally reduced over the non-acylated G (GDFRC-OH) and γ-p-coumaroylated G (GDFRC-pCA) monomeric compounds (Table 2, Supplementary Fig. S4). In addition, thioacidolysis determined significantly increased levels of 5-hydroxyguaiacyl (5H)-type monomeric compounds (5Hthio) in both OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls (Table 2; Supplementary Fig. S4), demonstrating the incorporation of unusual 5H units in lignins produced in the OsCAldOMT1-deficient rice cell walls.
2D NMR analysis of OsCAldOMT1-deficient rice lignins
For more in-depth lignin structural analysis, we performed 2D HSQC NMR analysis on non-acetylated and acetylated lignin samples prepared from rice culm tissues. The aromatic regions of the HSQC NMR spectra collected for wild-type rice lignins displayed intense signals from the canonical S and G lignin units along with those from the grass-specific tricin and pCA units; these aromatic signals were predictably shifted after acetylation (Fig. 4). In line with the chemical analysis data (Table 2), our HSQC data demonstrated that S lignins are largely depleted in the OsCAldOMT1-deficient rice lignins. Volume integration of the HSQC signals roughly estimated the decrease of S/G ratios to be 97% (from 1.22 to 0.04) and 94% (from 1.08 to 0.06) in the non-acetylated OsCAldOMT1-RNAi and OsCAldOMT1-TDNA lignin spectra, respectively; S lignin signals in OsCAldOMT1-deficient lignin spectra could be overestimated because signals from newly appearing 5H lignin units considerably overlap with S lignin signals27,28 (Fig. 4). In addition, we observed sharp reductions of the tricin signals in the OsCAldOMT1-deficient rice lignin spectra. Compared with the wild-type control spectra, tricin signals were reduced by more than 98% in both non-acetylated OsCAldOMT1-RNAi and OsCAldOMT1-TDNA lignin spectra (Fig. 4). These data firmly established that OsCAldOMT1 is requisite for normal formation of tricin lignin units in addition to S lignin units in rice cell walls.
Aromatic sub-regions of short range 1H–13C correlation (HSQC) NMR spectra of non-acetylated (upper) and acetylated (lower) samples of lignin-enriched CWRs prepared from culm tissues of OsCAldOMT1-RNAi, OsCAldOMT1-TDNA and their wild-type controls (WT1 and WT2). Contour coloration matches that of the lignin substructure units shown. Volume-integration data for the major aromatic units are shown in the non-acetylated lignin spectra.
The aliphatic side-chain regions of the HSQC spectra further provide information on the distribution of inter-monomeric linkages in the lignin polymers. Typical lignin linkage signals from β–O–4 (I), β–5 (II), and β–β (III) units were clearly seen in all wild-type and OsCAldOMT1-deficient lignin spectra (Fig. 5). Volume-integration analysis of these signals suggested that the proportions of β–O–4 units decreased, whereas the proportions of β–5 units increased in both OsCAldOMT1-RNAi and OsCAldOMT1-TDNA lignins (Fig. 5). In addition, new signals from the cyclic β–O–4 (benzodioxane) units (IV), which were not detected in the wild-type lignin spectra, were clearly visible in both the OsCAldOMT1-deficient rice lignin spectra [Cα–Hα correlations from IV (IVα) at δC/δH ~76/~5.0; Cβ–Hβ correlations (IVβ) from IV at δC/δH ~78/~4.1], accounting for 7% and 9% of the total detected inter-monomeric linkage signals (I, II, III, and IV) in the spectra of non-acetylated OsCAldOMT1-RNAi and OsCAldOMT1-TDNA lignins, respectively (Fig. 5). After acetylation, the benzodioxane signals predictably shifted [Cα–Hα correlations (IVα) at δC/δH ~76/~4.9; Cβ–Hβ correlations (IVβ) at δC/δH ~75/~4.2] (Fig. 5) and perfectly matched with the chemical shift data previously reported for several benzodioxane-containing lignin polymers57,58,59. This result indicates the incorporation of atypical catechol-type monomers such as 5-hydroxyconiferyl alcohol and/or selgin in lignification of the OsCAldOMT1-deficient rice lignins, as further discussed below (Supplementary Fig. S5).
Aliphatic sub-regions of short range 1H–13C correlation (HSQC) NMR spectra of non-acetylated (upper) and acetylated (lower) samples of lignin-enriched CWRs prepared from culm tissues of OsCAldOMT1-RNAi, OsCAldOMT1-TDNA and their wild-type controls. Contour coloration matches that of the inter-monomeric linkages shown (WT1 and WT2). Volume integration data for the major units with their characteristic inter-monomeric linkages are shown in the non-acetylated lignin spectra.
Enzymatic saccharification efficiency of OsCAldOMT1-deficient rice cell walls
We previously demonstrated that OsCAldOMT1-RNAi displayed enhanced biomass saccharification efficiency23. Here, we also evaluated the saccharification performance of OsCAldOMT1-TDNA together with OsCAldOMT1-RNAi under identical conditions. Consequently, we affirmed both the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls displayed similarly enhanced enzymatic saccharification efficiency over their wild-type controls (Table 2); compared with the wild-type controls, the enhancement in glucose yields from the OsCAldOMT1-RNAi and OsCAldOMT1-TDNA cell walls after 24 h incubation was similarly around 60%.
Discussion
In line with our previous study23, the two OsCAldOMT1-deficient rice lines analyzed in this study, i.e., OsCAldOMT1-RNAi and OsCAldOMT1-TDNA, both produced lignins largely depleted in S units and partially incorporated with atypical 5H units, as clearly demonstrated through in-depth cell wall structural analyses using chemical and NMR methods (Table 2, Figs 4 and 5). These results collectively support the notion that OsCAldOMT1 is a predominant CAldOMT functioning in S lignin biosynthesis in rice. Our analysis by DFRC further showed that both non-acylated and acylated S-type lignin degradation products (SDFRC-OH and SDFRC-pCA) were proportionally reduced with relative increases in the corresponding G-type products (GDFRC-OH and GDFRC-pCA) (Table 2, Supplementary Fig. S4). Recently, the existence of an alternative lignin biosynthetic pathway(s) that specifically leads to the production of grass-specific γ-p-coumaroylated lignin units has been suggested through the analysis of transgenic grass plants deficient in common lignin biosynthesis genes. For example, in rice, mutations of the CAld5H and C3′H genes, key regulators of the H/G/S lignin unit composition in eudicots5,6,7 (Fig. 1), predictably altered the H/G/S unit composition in the common non-acylated lignin units but exerted no detectable effects on the H/G/S unit composition in grass-specific γ-p-coumaroylated lignin units47,49. On the other hand, our current DFRC data suggest that OsCAldOMT1 may function in generating both non-acylated and γ-p-coumaroylated lignin units (Table 2, Supplementary Fig. S4).
Importantly, our histochemical and 2D NMR analysis firmly established that, along with S lignins, lignin-bound tricin was drastically reduced in the OsCAldOMT1-deficient rice cell walls (Figs 3 and 4), demonstrating the crucial role of OsCAldOMT1 in the biosynthesis of not only the sinapyl alcohol but also the tricin monomers for lignification; our NMR data suggested that lignin-integrated tricin units were nearly absent in the OsCAldOMT1-deficient rice cell walls (Fig. 4). Given that around 80% of tricin produced in typical grass straws can be lignin-bound14 and that we also observed substantial reductions of extractable tricin-O-conjugates in OsCAldOMT1-TDNA39, it is likely that OsCAldOMT1 plays a major role in the overall production of tricin-related metabolites in rice, including extractable tricin-O-conjugates and cell-wall-bound tricin-lignins.
Because lignin polymerization is a chemical process in which plants may incorporate any available phenolic compounds that can undergo phenoxy radical formation by laccases and/or peroxidases in lignifying cell walls, truncation of the monolignol biosynthetic pathway often results in the incorporation of pathway intermediates or their derivatives as non-canonical lignin monomers8,60,61; examples include the incorporation of caffeyl alcohol in a CCoAOMT-deficient plant62, ferulic acid in CCR-deficient plants63, p-hydroxycinnamaldehydes in CAD-deficient plants50,57 and, as we have demonstrated in this and previous studies with rice23, 5-hydroxyconiferyl alcohol in CAldOMT-deficient plants25,26,27. A similar phenomenon has also been observed when the tricin pathway is disrupted in grass species containing tricin-lignins; rice mutants deficient in OsFNSII18 and OsA3′H/C5′H19 produced lignins incorporating naringenin and apigenin, respectively, as non-canonical lignin monomers instead of the canonical tricin monomer.
In our OsCAldOMT1-deficient rice, the incorporation of 5-hydroxyconiferyl alcohol was evident by the appearance of the 5H-type monomeric compounds in the thioacidolysis-derived degradation products23 (Table 2, Supplementary Fig. S4). In addition, our 2D HSQC NMR analysis detected diagnostic signals from the benzodioxane linkages (Fig. 5), which can be uniquely derived from β–O–4-type radical coupling of a monolignol with 5-hydroxyguaiacyl end-groups (themselves derived from 5-hydroxyconiferyl alcohol incorporation) followed by internal trapping of quinone methide intermediates by the o-hydroxyl group64 (Supplementary Fig. S5). Such benzodioxanes, however, can be similarly derived from lignification of various types of o-diphenolic monomers58,65,66,67 and it is possible that analogous tricin pathway intermediates with o-diphenols such as selgin participate in lignification and produce benzodioxanes in CAldOMT-deficient grass lignins (Supplementary Fig. S5). This, however, is not likely for the OsCAldOMT1-deficient rice tested in this study because detection of any reasonable flavonoid signals failed, other than the small residual tricin signals in the HSQC NMR spectra of the OsCAldOMT1-deficient lignins (Fig. 4). Thus, it is likely that the benzodioxane units detected in the OsCAldOMT1-deficient lignins were mostly from the polymerization of 5-hydroxyconiferyl alcohol, rather than selgin derived from the tricin pathway (Fig. 1). The result was somewhat surprising because our previous metabolite analysis determined that selgin substantially over-accumulated in the vegetative tissues of OsCAldOMT1-TDNA39. It is currently unknown why selgin from the tricin pathway is not incorporated into lignins, whereas 5-hydroxyconiferyl alcohol from the monolignol pathway is readily incorporated in our OsCAldOMT1-deficient rice.
Although this study established that OsCAldOMT1 has comparable in vitro catalytic abilities towards 5-hydroxyconiferaldehyde and selgin, the potential in vivo substrates in the monolignol and tricin biosynthetic pathways, respectively (Fig. 2), such dual catalytic activity of grass CAldOMTs to catalyze both monolignol and flavonoid substrates has been documented. Prior to this work, OsCAldOMT1 was shown to catalyze O-methylation of not only monolignol-associated substrates23 but also 3′-O-methylation of a wide range of flavonoids, such as eriodictyol, luteolin, dihydroquercetin, quercetin, and rhamnetin68,69, as well as sequential 3′/5′-O-methylation of tricetin and/or myricetin39,70 under in vitro conditions. Likewise, other grass CAldOMT homologs including those in wheat71,72, maize73, barley73, and sorghum40 (Supplementary Fig. S1), have been reported to display in vitro OMT activities toward various flavone substrates in addition to monolignol-associated substrates.
In planta evidence for the involvement of CAldOMTs in tricin-lignin biosynthesis has also been provided in other grass species; cell wall analytical data recently reported for maize bm336 and sorghum bm1240 mutants harboring defects in their primary CAldOMT genes indicated that both S and tricin lignin units were concomitantly depleted in their lignins when compared to the wild-type controls, suggesting that, similarly to in rice, CAldOMT plays a key role in both the S lignin and tricin biosynthesis. Given also the conserved CAldOMT sequences (Supplementary Fig. S1) and the wide occurrence of tricin-lignins among grasses14, it is plausible that the utilization of bifunctional CAldOMT that generates tricin-lignins is a common feature of grasses and therefore modification of lignin and flavonoid content/composition in grass biomass may be achieved by manipulating a single CAldOMT target gene. In addition, as we have confirmed with rice (Table 2)23, CAldOMT-deficiency has been shown to positively impact the efficiency of biomass saccharification in many grass crops31,33,35,36,37,38. Collectively, CAldOMT holds promise as a potent bioengineering target for manipulating grass biomass structure and properties for improved biorefinery applications.
Methods
Bioinformatics
A phylogenetic tree was constructed by the neighbor-joining method using MEGA774. Bootstrapping with 1,000 replications was performed. Microarray-based gene expression profiles were retrieved from the Rice Expression Profile Database46.
CAldOMT enzyme assay
Expression of recombinant OsCAldOMT1 in Escherichia coli and subsequent purification of the enzyme were carried out as described previously22,23. For enzymatic assays, selgin was synthesized according to an analogous synthetic route previously described for tricin13 and synthetic protocols for 5-hydroxyconiferaldehyde are described in Sakakibara et al.75. The typical reaction mixture (200 μl) contained 149 μl of 50 mM Tris-HCl (pH 7.5), 8 μl of 5 mM S-adenosyl-l-methionine, 20 μl of 5-hydroxyconiferaldehyde or selgin substrate solution in DMSO and 5 μg (23 μl) of recombinant enzyme in 50 mM Tris-HCl (pH 7.5)23. Three independent reactions for 8 different substrate concentrations ranging from 10 to 200 µM were carried out. After incubation for 20 min, the reaction was terminated by adding 200 μl of 2 M HCl and 10 μl of 1 mg ml−1 3,4,5-trimethoxycinnamic acid in DMSO as an internal standard. The reaction mixture was extracted with ethyl acetate, evaporated and re-dissolved in 50 μl of methanol. After filtration through a PVDF membrane with a 0.45 μm pore size (Ultrafree-MC-HV; Merck Millipore, Tullagreen, Co. Cork, Ireland), the reaction mixtures were analyzed by Shimadzu LCMS-2020 (Shimadzu, Kyoto, Japan) with the following conditions: injection volume, 3 μl; column, Gemini 5 μm C18 110 Å 150 × 2 mm (Phenomenex, USA); solvents, 0.1% (v/v) formate/water (solvent A) and 0.1% (v/v) formate/acetonitrile (solvent B); solvent gradient protocol, linear gradient of 90% solvent A and 10% solvent B to 50% solvent A and solvent B for 20 min; UV detection, at 254 nm; MS ionization, ESI (+/−); MS interface, 350 °C; MS desolvation line and heat block, 200 °C; MS nebulizer gas, N2 at 1.5 l min−1. Compound identification was carried out based on comparisons of retention time and MS fragmentation pattern with authentic standards. Calibration curves for quantification were constructed using authentic standards and 254 nm UV absorbance for peak detection and area calculation. The Michaelis-Menten curves for the determination of Vmax and Km values were obtained with the GraphPad Prism ver. 8.1.2 program (GraphPad Software Inc, San Diego, CA).
Plant materials
OsCAldOMT1-RNAi plants (cv. Nipponbare; T3 generation) were derived from Koshiba et al.23. OsCAldOMT1-TDNA plants (cv. Hwayoung; accession number: PFG_2B-50240) were originally from the Crop Biotech Institute at Kyung Hee University76, and the homozygous mutant and near-isogenic wild-type lines were isolated as described previously39. Rice seeds were surface sterilized, germinated and grown in a phytotron with a photoperiod of 12 h light (~30 °C) and 12 h dark (~24 °C). Mature plants (~45 days after heading) were phenotypically characterized, harvested and dried at ~27 °C for 30 days prior to cell wall analysis18,23.
Histochemistry
Fresh hand-cut specimens were excised from rice culms at the heading stage, fixed, sectioned and stained by phloroglucinol-HCl or vanillin-HCl as described previously18. The stained sections were visualized under an Olympus BX51 microscope (Olympus Optical, Tokyo, Japan).
Cell wall sample preparations
Extractive-free cell wall residues (CWRs) for chemical analysis were prepared from culm samples of matured rice plants77. For NMR analysis, CWRs (~300 mg) were further ball-milled using the Planetary Micro Mill Pulverisette 7 (Fritsch Industrialist, Idar-Oberstein, Germany) equipped with ZrO2 vessels containing ZrO2 ball bearings (600 rpm, 12 cycles of 10 min ball-milling with 5 min intervals), followed by digestion using crude cellulases (Cellulysin; Calbiochem, La Jolla, CA, USA) to give lignin-enriched CWRs59. Aliquots of the lignin-enriched CWRs (~15 mg) were dissolved in DMSO-d6/pyridine-d5 (4:1, v/v) and subjected to 2D NMR analysis. In parallel, the lignin-enriched CWRs (~15 mg) were further acetylated in DMSO/N-methylimidazole/acetic anhydride (2:1:1, v/v/v)59,78 and dissolved in chloroform-d for additional 2D NMR data analysis.
Chemical analyses
Klason lignin79, sugar analysis18, quantitation of cell-wall-bound pCA and FA released by mild-alkaline hydrolysis80, analytical thioacidolysis77,81, and DFRC analysis47,82 were performed in accordance with the methods described in the literature.
2D HSQC NMR analysis
NMR spectra were acquired on a Bruker Biospin Avance III 800 system (800 MHz, Bruker Biospin, Billerica, MA, USA) equipped with a cryogenically-cooled 5-mm TCI gradient probe. Adiabatic 2D HSQC NMR experiments were carried out using a standard Bruker implementation (hsqcetgpsp.3, Bruker Biospin, Billerica, MA, USA) with parameters described in the literature62,83, and data processing and analysis were carried out as described previously18,59. The central solvent peaks were used as internal references for chemical shift calibration (δC/δH: DMSO, 39.5/2.49 ppm; chloroform, 77.0/7.26 ppm). For volume integration of lignin signals, C2–H2 correlations from G units, C2–H2/C6–H6 correlations from S units and C2′–H2′/C6′–H6′ correlations from tricin units were used. Signals from S and tricin units were logically halved. The percentages in Fig. 4 were calculated based on G + S = 100. Volume integrations of lignin intermonomeric linkages were carried out using Cα–Hα contours from I, II, III and IV units. Signals from III were logically halved. The percentages shown in Fig. 5 were expressed based on I + II + III + IV = 100.
Enzymatic saccharification
Enzymatic saccharification was carried out essentially in accordance with Hattori et al.84 and Lam et al.18. Briefly, destarched CWRs were digested by a cocktail of commercial cellulolytic enzymes that contain Celluclast 1.5 L, Novozyme 188 and Ultraflo L (Novozymes, Bagsværd, Denmark) dissolved in 50 mM sodium citrate (pH 4.8). After 6 and 24 h of cell wall digestion, the glucose yield was measured using Glucose C-II test kit (Wako Pure Chemical Industries, Osaka, Japan), according to the manufacturer’s instruction.
Accession numbers
Sequence data of rice OsCAldOMT1 can be found in the EMBL/GenBank data libraries under accession number Q6ZD89. Accession numbers for the sequences used in the phylogenetic analysis (Supplementary Fig. S1) and in silico gene expression analysis (Supplementary Fig. S2) are listed in Supplementary Information.
Data Availability
All data necessary to evaluate the conclusions in this study are included in the published paper and its Supplementary Information file. Additional data, if required, will be made available by the corresponding authors upon request.
References
Tye, Y. Y., Lee, K. T., Abdullah, W. N. W. & Leh, C. P. The world availability of non-wood lignocellulosic biomass for the production of cellulosic ethanol and potential pretreatments for the enhancement of enzymatic saccharification. Renew. Sust. Energ. Rev. 60, 155–172 (2016).
Umezawa, T. Lignin modification in planta for valorization. Phytochem. Rev. 17, 1305–1327 (2018).
Cesarino, I. et al. Building the wall: recent advances in understanding lignin metabolism in grasses. Acta Physiol. Plant 38, 269 (2016).
Bhatia, R., Gallagher, J. A., Gomez, L. D. & Bosch, M. Genetic engineering of grass cell wall polysaccharides for biorefining. Plant Biotechnol. J. 15, 1071–1092 (2017).
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003).
Umezawa, T. The cinnamate/monolignol pathway. Phytochem. Rev. 9, 1–17 (2010).
Bonawitz, N. D. & Chapple, C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 (2010).
Mottiar, Y., Vanholme, R., Boerjan, W., Ralph, J. & Mansfield, S. D. Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200 (2016).
Ragauskas, A. J. et al. Lignin valorization: improving lignin processing in the biorefinery. Science 344, 1246843 (2014).
Rinaldi, R. et al. Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew. Chem. Int. Edit. 55, 8164–8215 (2016).
Halpin, C. Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. (2019).
del Río, J. C. et al. Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60, 5922–5935 (2012).
Lan, W. et al. Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 167, 1284–1295 (2015).
Lan, W. et al. Tricin-lignins: occurrence and quantitation of tricin in relation to phylogeny. Plant J. 88, 1046–1057 (2016).
Ibrahim, R. K. & Anzellotti, D. The enzymatic basis of flavonoid biodiversity. Rec. Adv. Phytochem. 37, 1–36 (2003).
Lepiniec, L. et al. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430 (2006).
Ralph, J. Hydroxycinnamates in lignification. Phytochem. Rev. 9, 65–83 (2010).
Lam, P. Y. et al. Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol. 174, 972–985 (2017).
Lam, P. Y. et al. Recruitment of specific flavonoid B-ring hydroxylases for two independent biosynthesis pathways of flavone-derived metabolites in grasses. New Phytol. 223, 204–219 (2019).
Osakabe, K. et al. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96, 8955–8960 (1999).
Li, L., Popko, J. L., Umezawa, T. & Chiang, V. L. 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275, 6537–6545 (2000).
Nakatsubo, T. et al. Roles of 5-hydroxyconiferylaldehyde and caffeoyl CoA O-methyltransferases in monolignol biosynthesis in Carthamus tinctorius. Cell. Chem. Technol. 41, 511–520 (2007).
Koshiba, T. et al. Characterization of 5-hydroxyconiferaldehyde O-methyltransferase in Oryza sativa. Plant Biotechnol. 30, 157–167 (2013).
Nakatsubo, T. et al. At5g54160 gene encodes Arabidopsis thaliana 5-hydroxyconiferaldehyde O-methyltransferase. J. Wood Sci. 54, 312–317 (2008).
Jouanin, L. et al. Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123, 1363–1374 (2000).
Guo, D., Chen, F., Inoue, K., Blount, J. W. & Dixon, R. A. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88 (2001).
Ralph, J. et al. NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into lignins of O-methyltransferase-deficient poplars. J. Agric. Food Chem. 49, 86–91 (2001).
Vanholme, R. et al. Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J. 64, 885–897 (2010).
Weng, J. K., Mo, H. & Chapple, C. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64, 898–911 (2010).
Sattler, S. E., Funnell-Harris, D. L. & Pedersen, J. F. Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci. 178, 229–238 (2010).
Sattler, S. E. et al. Identification and characterization of four missense mutations in brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenergy Res. 5, 855–865 (2012).
Tu, Y. et al. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22, 3357–3373 (2010).
Fu, C. et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. USA 108, 3803–3808 (2011).
Jung, J. H., Fouad, W. M., Vermerris, W., Gallo, M. & Altpeter, F. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 10, 1067–1076 (2012).
Ho-Yue-Kuang, S. et al. Mutation in Brachypodium caffeic acid O-methyltransferase 6 alters stem and grain lignins and improves straw saccharification without deteriorating grain quality. J. Exp. Bot. 67, 227–237 (2015).
Fornalé, S. et al. Changes In cell wall polymers and degradability in maize mutants lacking 3′- and 5′-O-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol. 58, 240–255 (2016).
Kannan, B., Jung, J. H., Moxley, G. W., Lee, S. M. & Altpeter, F. TALEN mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16, 856–866 (2017).
Daly, P. et al. RNAi-suppression of barley caffeic acid O-methyltransferase modifies lignin despite redundancy in the gene family. Plant Biotechnol. J. 17, 594–607 (2019).
Lam, P. Y., Liu, H. & Lo, C. Completion of tricin biosynthesis pathway in rice: Cytochrome P450 75B4 is a novel chrysoeriol 5′-hydroxylase. Plant Physiol. 175, 1527–1536 (2015).
Eudes, A. et al. SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PloS One 12, e0178160 (2017).
Ma, Q. H. & Xu, Y. Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie 90, 515–524 (2008).
Goujon, T. et al. A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51, 973–989 (2003).
Do, C. T. et al. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129 (2007).
Moinuddin, S. G. et al. Insights into lignin primary structure and deconstruction from Arabidopsis thaliana COMT (caffeic acid O-methyl transferase) mutant Atomt1. Org. Biomol. Chem. 8, 3928–3946 (2010).
Pinçon, G. et al. Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57, 1167–1176 (2001).
Sato, Y. et al. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, D1206–1213 (2012).
Takeda, Y. et al. Downregulation of p-COUMAROYL ESTER 3-HYDROXYLASE in rice leads to altered cell wall structures and improves biomass saccharification. Plant J. 95, 796–811 (2018).
Takeda, Y. et al. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 246, 337–349 (2017).
Takeda, Y. et al. Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J. 97, 543–554 (2019).
Koshiba, T. et al. CAD2 deficiency causes both brown midrib and gold hull and internode phenotypes in Oryza sativa L. cv. Nipponbare. Plant Biotechnol. 30, 365–373 (2013).
Withers, S. et al. Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 287, 8347–8355 (2012).
Shih, C. H. et al. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228, 1043–1054 (2008).
Hong, L. et al. A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta 236, 141–151 (2012).
Lam, P. Y., Zhu, F. Y., Chan, W. L., Liu, H. & Lo, C. Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol. 165, 1315–1327 (2014).
Lapierre, C., Monties, B. & Rolando, C. Preparative thioacidolysis of spruce lignin: isolation and identification of main monomeric products. Holzforschung 40, 47–50 (1986).
Lu, F. & Ralph, J. The DFRC method for lignin analysis. 7. Behavior of cinnamyl end groups. J. Agric. Food Chem. 47, 1981–1987 (1999).
Ralph, J. et al. NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 95, 12803–12808 (1998).
Chen, F., Tobimatsu, Y., Havkin-Frenkel, D., Dixon, R. A. & Ralph, J. A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA. 109, 1772–1777 (2012).
Tobimatsu, Y. et al. Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25, 2587–2600 (2013).
Ralph, J. et al. Syntax of referencing in Recent Advances in Polyphenol Research, Vol 1. (eds Daayf, F. and Lattanzio, V.) 36–66 (Wiley-Blackwell Publishing, 2008).
Tobimatsu, Y. & Schuetz, M. Lignin polymerization: how do plants manage the chemistry so well? Curr. Opin. Biotechnol. 56, 75–81 (2019).
Wagner, A. et al. CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J. 67, 119–129 (2011).
Ralph, J. et al. Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J. 53, 368–379 (2008).
Marita, J. M., Vermerris, W., Ralph, J. & Hatfield, R. D. Variations in the cell wall composition of maize brown midrib mutants. J. Agric. Food Chem. 51, 1313–1321 (2003).
Elumalai, S., Tobimatsu, Y., Grabber, J. H., Pan, X. & Ralph, J. Epigallocatechin gallate incorporation into lignin enhances the alkaline delignification and enzymatic saccharification of cell walls. Biotechnol. Biofuels 5, 59 (2012).
Tobimatsu, Y. et al. Hydroxycinnamate conjugates as potential monolignol replacements: in vitro lignification and cell wall studies with rosmarinic acid. ChemSusChem 5, 676–686 (2012).
del Río, J. C., Rencoret, J., Gutiérrez, A., Kim, H. & Ralph, J. Hydroxystilbenes are monomers in palm fruit endocarp lignins. Plant Physiol. 174, 2072–2082 (2017).
Kim, B. G., Lee, Y., Hur, H. G., Lim, Y. & Ahn, J. H. Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67, 387–394 (2006).
Lin, F. et al. Cloning and functional analysis of caffeic acid 3-O-methyltransferase from rice (Oryza sativa). J. Pestic. Sci. 31, 47–53 (2006).
Zhou, J. M., Fukushi, Y., Wang, X. F. & Ibrahim, R. K. Characterization of a novel flavone O-methyltransferase gene in rice. Nat. Prod. Commun. 1, 981–984 (2006).
Zhou, J. M., Gold, N. D., Martin, V. J., Wollenweber, E. & Ibrahim, R. K. Sequential O-methylation of tricetin by a single gene product in wheat. Biochim. Biophys. Acta Gen. Subj. 1760, 1115–1124 (2006).
Zhou, J. M. et al. Structure-function relationships of wheat flavone O-methyltransferase: Homology modeling and site-directed mutagenesis. BMC Plant Biol. 10, 156 (2010).
Zhou, J. M., Fukushi, Y., Wollenweber, E. & Ibrahim, R. K. Characterization of two O-methyltransferase-like genes in barley and maize. Pharm. Biol. 46, 26–34 (2008).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Sakakibara, N. et al. Metabolic analysis of the cinnamate/monolignol pathway in Carthamus tinctorius seeds by a stable-isotope-dilution method. Org. Biomol. Chem. 5, 802–815 (2007).
An, S. et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133, 2040–2047 (2003).
Yamamura, M., Hattori, T., Suzuki, S., Shibata, D. & Umezawa, T. Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol. 29, 419–423 (2012).
Lu, F. & Ralph, J. Non-degradative dissolution and acetylation of ball-milled plant cell walls: high-resolution solution-state NMR. Plant J. 35, 535–544 (2003).
Hatfield, R. D., Jung, H. J. G., Ralph, J., Buxton, D. R. & Weimer, P. J. A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J. Sci. Food Agric. 65, 51–58 (1994).
Yamamura, M. et al. Occurrence of guaiacyl/p-hydroxyphenyl lignin in Arabidopsis thaliana T87 cells. Plant Biotechnol. 28, 1–8 (2011).
Yue, F., Lu, F., Sun, R.-C. & Ralph, J. Syntheses of lignin-derived thioacidolysis monomers and their uses as quantitation standards. J. Agric. Food Chem. 60, 922–928 (2012).
Karlen, S. D. et al. Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2, e1600393 (2016).
Mansfield, S. D., Kim, H., Lu, F. & Ralph, J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7, 1579–1589 (2012).
Hattori, T. et al. Rapid analysis of transgenic rice straw using near-infrared spectroscopy. Plant Biotechnol. 29, 359–366 (2012).
Acknowledgements
We thank Ms. Keiko Tsuchida and Ms. Megumi Ozaki for assisting in the analysis of rice cell walls, Mr. Keisuke Kobayashi for assisting in the recombinant enzyme assay, Dr. Hironori Kaji and Ms. Ayaka Maeno for their assistance in NMR analysis, and Dr. Arata Yoshinaga and Dr. Keiji Takabe for assistance with the histochemical analysis and helpful suggestions. This work was supported in part by grants from the Japan Science and Technology Agency/Japan International Cooperation Agency (Science and Technology Research Partnership for Sustainable Development, SATREPS), the Japan Society for the Promotion of Science (grant nos. KAKENHI #16K14958 and #16H06198), RISH Kyoto University (grant no. Mission-linked Research Funding #2016-5-2-1), and Research Grants Council of Hong Kong, China (grant nos. GRF17113217 and GRF17126918). P.Y.L. and Y.Ta. acknowledge the JSPS fellowship programs (program nos. #17F17103 and #17J0965416). W.L. and J.R. were funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494 and DE-SC0018409). A part of this study was conducted using the facilities at the DASH/FBAS (Research Institute for Sustainable Humanosphere, Kyoto University) and the NMR spectrometer at JURC (Institute for Chemical Research, Kyoto University).
Author information
Authors and Affiliations
Contributions
P.Y.L., Y.To., N.M., S.S., W.L., Y.Ta. and M.Y. performed experiments, P.Y.L., Y.To., S.S., M.S., J.R., C.L. and T.U. conceived research, analyzed data, and P.Y.L., Y.To. and T.U. wrote the manuscript with help from all the others.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lam, P.Y., Tobimatsu, Y., Matsumoto, N. et al. OsCAldOMT1 is a bifunctional O-methyltransferase involved in the biosynthesis of tricin-lignins in rice cell walls. Sci Rep 9, 11597 (2019). https://doi.org/10.1038/s41598-019-47957-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47957-0
This article is cited by
-
Strong culm: a crucial trait for developing next-generation climate-resilient rice lines
Physiology and Molecular Biology of Plants (2024)
-
Development of an in vitro regeneration system from immature inflorescences and CRISPR/Cas9-mediated gene editing in sudangrass
Journal of Genetic Engineering and Biotechnology (2023)
-
Flavonoids in major cereal grasses: distribution, functions, biosynthesis, and applications
Phytochemistry Reviews (2023)
-
Nitrogen deficiency results in changes to cell wall composition of sorghum seedlings
Scientific Reports (2021)
-
Altered lignocellulose chemical structure and molecular assembly in CINNAMYL ALCOHOL DEHYDROGENASE-deficient rice
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.