Abstract
Background:
We have recently identified down-regulated microRNAs including miR-145 and miR-133a in bladder cancer (BC). The aim of this study is to determine the genes targeted by miR-145, which is the most down-regulated microRNA in BC.
Methods:
We focused on fascin homologue 1 (FSCN1) from the gene expression profile in miR-145 transfectant. The luciferase assay was used to confirm the actual binding sites of FSCN1 mRNA. Cell viability was evaluated by cell growth, wound-healing, and matrigel invasion assays. BC specimens were subjected to immunohistochemistry of FSCN1 and in situ hybridisation of miR-145.
Results:
The miR-133a as well as miR-145 had the target sequence of FSCN1 mRNA by the database search, and both microRNAs repressed the mRNA and protein expression of FSCN1. The luciferase assay revealed that miR-145 and miR-133a were directly bound to FSCN1 mRNA. Cell viability was significantly inhibited in miR-145, miR-133a, and si-FSCN1 transfectants. In situ hybridisation revealed that miR-145 expression was markedly repressed in the tumour lesion in which FSCN1 was strongly stained. The immunohistochemical score of FSCN1 in invasive BC (n=46) was significantly higher than in non-invasive BC (n=20) (P=0.0055).
Conclusion:
Tumour suppressive miR-145 and miR-133a directly control oncogenic FSCN1 in BC.
Similar content being viewed by others
Main
Bladder cancer (BC) is the fifth most common cancer in the United States and the second most common cancer of the genitourinary tract (Parkin et al, 2005; Jemal et al, 2008). In Japan, the age-standardised mortality rate of BC has increased slightly since 1993 (Qiu et al, 2009). Currently, the standard diagnostic method depends on the use of invasive urethro-cystoscopy. Bladder tumour antigen and nuclear matrix protein-22 are available as urine markers for BC diagnostic tools. However, they are not widely used because of their low sensitivity and specificity for distinguishing BC from non-malignant diseases (van Rhijn et al, 2005). In the treatment of BC, morphologically similar tumours can behave differently, and it is currently not possible to identify patients who will experience tumour recurrence or disease progression (Kwak et al, 2004). Therefore, a new diagnostic method and treatment based on BC biology are desired.
MicroRNAs are an abundant class of small non-coding RNAs of about 22 nucleotides in length that function as negative regulators of gene expression through antisense complimentarily to specific messenger RNAs (Lagos-Quintana et al, 2001). Although their biological functions remain largely unknown, recent studies suggest that microRNAs contribute to the development of various cancers (Schickel et al, 2008). The miR-145 and miR-133a/b have been commonly identified as down-regulated in the microRNA expression signatures of various human malignancies: head and neck carcinoma (Wang et al, 2008; Wong et al, 2008a; Childs et al, 2009), pancreatic ductal adenocarcinoma (Szafranska et al, 2007), lung cancer (Liu et al, 2009), breast cancer (Sempere et al, 2007; Wang et al, 2009a), gastric cancer (Takagi et al, 2009), colorectal cancer (Bandrés et al, 2006; Slaby et al, 2007; Schepeler et al, 2008; Wang et al, 2009b), prostate cancer (Ozen et al, 2008; Tong et al, 2009), and BC (Friedman et al, 2009; Lin et al, 2009a). In our microRNA screening test of BC, we identified a subset of seven differentially down-regulated microRNAs (miR-145, miR-133a, miR-133b, miR-30a-3p, miR-195, miR-125b, and miR-199a*) among the 156 microRNAs examined, and miR-145 was the most down-regulated one of all (Ichimi et al, 2009). These studies strongly suggest that low expression levels of miR-145 and miR-133a/b may contribute to pathogenesis and progression of human malignancies. Moreover, functional analyses of target genes, which are repressed by these microRNAs, are crucial to elucidate the mechanisms of cancer development. In this study, we performed an oligo-microarray analysis of miR-145-transfected BC cell lines in comparison with their parental cell lines for genome-wide screening of target genes silenced by miR-145 in BC, and we found that fascin homologue 1 (FSCN1) was the most down-regulated one among the genes.
FSCN1 is an actin-binding protein required for the formation of actin-based cell-surface protrusions and cytoplasmic bundles of microfilaments (Hashimoto et al, 2005). FSCN1 activity is essential to filopodial dynamics, and it has been proposed that fascin imparts rigidity to the forming filopodia to efficiently push the membrane forwards (Vignjevic et al, 2006). Cells with prominent cytoplasmic protrusions and actively migrating cells express high levels of FSCN1, whereas this protein is undetectable in most normal epithelial cells (Pelosi et al, 2003). Over-expression of FSCN1 in a variety of tumours such as lung (Pelosi et al, 2003), prostate (Darnel et al, 2009), oesophageal (Zhang et al, 2006), breast (Grothey et al, 2000), colon (Jawhari et al, 2003), pancreas (Maitra et al, 2003), ovary (Lin et al, 2009b), and skin cancers (Goncharuk et al, 2002) usually correlates with high-grade, extensive invasion, distant metastasis, and poor prognosis. However, little is known about the function of FSCN1 in BC, and it is not known whether FCSN1 expression is regulated by specific microRNAs.
We hypothesised that miR-145 and miR-133a/b directly regulate FSCN1 and that FSCN1 has oncogenic activity in BC. We used a luciferase reporter assay to determine whether FSCN1 actually has sites targeted by miR-145 and miR-133a. To investigate the functional roles of FSCN1 in BC, we performed a loss-of-function study using BC cell lines. Furthermore, we evaluated FSCN1 protein expression in clinical BC specimens by immunohistochemistry.
Materials and methods
Clinical samples and cell culture
The tissue specimens were from 66 BC patients who had undergone cystectomy or transurethral resection of bladder tumours at Kagoshima University Hospital between 2001 and 2005. The patient's background and clinico-pathological characteristics are summarised in Table 1. These samples were staged according to the American Joint Committee on Cancer-Union Internationale Contre le Cancer tumour-node-metastasis classification and histologically graded (Sobin and Wittekind, 2002). Normal bladder epithelia (N1 and N2) were derived from patients with non-cancerous disease and were used as the controls. Our study was approved by the Bioethics Committee of Kagoshima University; written prior informed consent and approval were given by the patients. We used three human BC cell lines; BOY was established in our laboratory from an Asian male patient aged 66 years, who had a diagnosis of stage III BC with lung metastasis (Ichimi et al, 2009); T24 was obtained from American Type Culture Collection; and KK47 was established in Kanazawa University and kindly provided. These cell lines were maintained in a minimum essential medium (MEM) supplemented with 10% foetal bovine serum in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Mature microRNA and siRNA transfection
As earlier described (Ichimi et al, 2009), the transfection of BC cell lines was accomplished with RNAiMAX transfection reagent (Invitrogen, Carlisbad, CA, USA), Opti-MEM (Invitrogen) with 10 nM of mature microRNA molecules. For gain-of-function experiments, we used Pre-miR and negative-control microRNA (Applied Biosystems, Foster City, CA, USA), whereas FSCN1 siRNA (LU-019576-00, J-019576-07, J-019576-08; Thermo Fisher Scientific, Waltham, MA, USA) and negative-control siRNA (D-001810–10; Thermo Fisher Scientific) were used for loss-of-function experiments. Cells were seeded under the following conditions: 800 000 in a 10 cm dish for protein extraction, 3000 per well in a 96-well plate for XTT assay, 200 000 per well in a 6-well plate for the wound-healing assay, and 50 000 per well in a 24-well plate for the mRNA extraction, matrigel invasion assay, and luciferase assay.
Quantitative real-time RT–PCR
TaqMan probes and primers for FSCN1 (P/N: Hs00979631_g1; Applied Biosystems) were assay-on-demand gene expression products. All reactions were performed in duplicate and a negative-control lacking cDNA was included. Regarding the PCR conditions, we followed the manufacturer's protocol. Stem-loop RT–PCR (TaqMan MicroRNA Assays; Applied Biosystems) was used to quantitate microRNAs according to the earlier published conditions (Ichimi et al, 2009). For quantitative analysis of FSCN1 mRNA and the microRNAs, human 18s rRNA (P/N: Hs99999901_s1; Applied Biosystems) and RNU48 (P/N: 001006; Applied Biosystems), respectively, served as internal controls, and the delta–delta Ct methods to calculate the fold change. We used premium total RNA from normal human bladder (Clontech, Mountain View, CA, USA) as a reference.
Gene expression analysis of BC cell lines
Total RNA was extracted by using TRIzol (Invitrogen) according to the manufacturer's protocol. The integrity of the RNA was checked with an RNA 6000 Nano Assay kit and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Oligo-microarray Human 44K (Agilent Technologies) was used for expression profiling in miR-145-transfected BC cell lines (T24 and KK47) in comparison with miR-negative-control transfectant, as described earlier (Sugimoto et al, 2009). Briefly, hybridisation and washing steps were performed in accordance with the manufacturer's instructions. The arrays were scanned using a Packard GSI Lumonics ScanArray 4000 (Perkin Elmer, Boston, MA, USA). The data obtained were analysed by means of DNASIS array software (Hitachi Software Engineering), which converted the signal intensity for each spot into text format. The Log2 ratios of the median subtracted background intensity were analysed. Data from each microarray study were normalised by the global normalisation method.
Western blots
After 3 days of transfection, protein lysate (50 μg) was separated by NuPAGE on 4–12% bis–tris gel (Invitrogen) and transferred into a polyvinylidene fluoride membrane. Immunoblotting was carried out with diluted (1 : 100) monoclonal FSCN1 antibody (ab49815, Abcam, Cambridge, UK) and GAPDH antibody (MAB374; Chemicon, Temecula, CA, USA). The membrane was washed and then incubated with goat anti-mouse IgG (H+L)-HRP conjugate (Bio-Rad, Hercules, CA, USA). Specific complexes were visualised with an echochemiluminescence detection system (GE Healthcare, Little Chalfont, UK).
Cell growth, wound-healing, and matrigel invasion assays
Cell growth was determined by using an XTT assay (Roche Applied Sciences, Tokyo, Japan) that was performed according to the manufacturer's instructions. Cell migration activity was evaluated by wound-healing assay. Cells were plated in six-well dishes, and the cell monolayer was scraped using a micropipette tip. The initial gap length (0 h) and the residual gap length 24 h after wounding were calculated from Photomicrographs. A cell invasion assay was carried out using modified Boyden Chambers consisting of transwell-precoated matrigel membrane filter inserts with 8 μm pores in 24-well tissue culture plates (BD Biosciences, Bedfold, MA, USA). MEM containing 10% foetal bovine serum in the lower chamber served as the chemoattractant. All experiments were performed in triplicate.
Prediction of microRNA targets
To investigate the predicted target genes and their conserved sites in which the seed region of each microRNA binds, we used the TargetScan program (release 5.0, http://www.targetscan.org/). The sequences of the predicted mature microRNAs were confirmed by referring miRBase (release 13.0, http://microrna.sanger.ac.uk/).
Plasmid construction and dual-luciferase assay
MicroRNA target sequences were inserted between the XhoI–PmeI restriction sites in the 3′-UTR of the hRluc gene in psiCHECK-2 vector (C8021, Promega, Madison, WI, USA). BOY cells were transfected with 5 ng of vector, 10 nM of microRNAs, and 1 μl of Lipofectamine 2000 (Invitrogen) in a 100 μl Opti-MEM. The activities of firefly and Renilla luciferases in cell lysates were determined with a dual-luciferase assay system (Promega). Normalised data were calculated as the quotient of Renilla/firefly-luciferase activities.
Immunohistochemistry
The primary mouse monoclonal antibodies against FSCN1 (Abcam) were diluted by 1 : 200. The slides were treated with Biotinylated Anti-Mouse IgG (H+L) made in horse (Vector laboratories, Burlingame, CA, USA). Diaminobenzidine-hydrogen peroxide (Sigma-Aldrich, St Louis, MO, USA) was the chromogen, and the counterstaining was carried out with 0.5% haematoxylin. The positivity of endothelia and myofiblasts served as an inner positive control. The intensity of the staining was scored as negative (0), weak (1), moderate (2), or strong (3) (Ropponen et al, 1999). All staining scores are averages of duplicate experiments, and all samples were independently scored by two of us (TC and HE) who were blinded to the patient status.
In situ hybridisation of microRNA
In situ hybridisation was carried out according to the manufacturer's protocol for formalin-fixed, paraffin-embedded (FFPE) tissue (Kloosterman et al, 2006) on human BC specimens. DIG-labelled LNA oligo-nucleotides were purchased from EXIQON (Woburm, MA, USA) and used for overnight hybridisation at 52°C. The staining was carried out as described earlier. After deparaffinisation, the specimens were subjected to proteinase K (20 Ag per ml) digestion for 20 min. The post-fixed tissues were subsequently incubated overnight with the locked nucleic acid-modified probes. For the immunodetection, tissues were incubated overnight at 4°C in anti-DIG-AP FAB fragment (Roche Applied Sciences; 1/2000). The final visualisation was carried out with NBT/BCIP (Pierce, Rockford, IL, USA).
Statistical analysis
The relationship between two variables and the numerical values obtained by real-time RT–PCR was analysed using the Mann–Whitney U-test. The relationship between three variables and the numerical values was analysed using the Bonferroni-adjusted Mann–Whitney U-test. The analysis software was Expert StatView (version 4, SAS Institute Inc., Cary, NC, USA); for the comparison test among the three variables, a non-adjusted statistical level of significance of P<0.05 corresponds to a Bonferroni-adjusted level of P<0.0167.
Results
Gene expression profile identifying down-regulated genes in miR-145 transfectant
For genome-wide screening of target genes silenced by miR-145 in BC, we performed an oligo-microarray analysis of miR-145-transfected BC cell lines (T24 and KK47) in comparison with miR-negative-control transfectant. A total of 200 genes were generally down-regulated by >0.5-fold in miR-145 transfectants compared with the control. We focused on FSCN1 because it was listed as the top candidate in the expression profile (Table 2).
FSCN1 as a target of post-transcriptional repression by miR-145 and miR-133a
Among the T24 cell lines transfected with the six down-regulated microRNAs in our earlier study (Ichimi et al, 2009), the expression levels of FSCN1 mRNA and its protein were markedly decreased not only in miR-145, but also in miR-133a transfectants (Figure 1A and B). We performed a luciferase assay to determine whether FSCN1 mRNA actually has the target sites of these two microRNAs, as indicated by the TargetScan algorithm. We initially used the vector encoding full-length 3′-UTR of FSCN1 mRNA (position 51–1180), and the luminescence intensity was significantly decreased in miR-145 and miR-133a transfectants (Figure 2A). Furthermore, to determine the specific sites targeted by the two microRNAs, we constructed vectors covering four conserved sites for miR-145 and one site for miR-133a (Figure 2B). The luminescence intensity was significantly decreased at the two sites targeted by miR-145 (positions 377–383 and 1140–1146) and one site targeted by miR-133a (position 745–751) (Figure 2B). In addition, we constructed three mutated vectors in which the specific sites targeted by the microRNAs were deleted, and the luminescence intensity was not decreased at all by miR-145 and miR-133a (Figure 2C). We did not examine miR-133b because it was considered to function similarly to miR-133a; these microRNAs have very similar sequences (miR-133a: UUGGUCCCCUUCAACCAGCUGU, miR-133b: UUGGUCCCCUUCAACCAGCUA) and have a common sequence for binding to FSCN1 mRNA (UUGGUC) (Figure 2B).
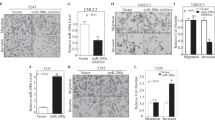
Regulation of FSCN1 expression in the down-regulated microRNA transfectants (T24). (A) FSCN1 mRNA expression after 24 h transfection with 10 nM of microRNAs (miR-145, miR-30a-3p, miR-133a, miR-195, miR-125b, and miR-199a*). FSCN1 mRNA expression was repressed in miR-145 and miR-133a transfectants. (B) FSCN1 protein expression after 72 h transfection of microRNAs. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The protein expression level of FSCN1 was also repressed in the transfectants.
miR-145- and miR-133a-binding sites in FSCN1 3′-UTR. (A) A luciferase assay using the vector encoding full length of FSCN1 3′-UTR (position 51–1180). BOY cells were transfected with 5 ng vector and 10 nM microRNAs. The Renilla luciferase values were normalised by firefly-luciferase values. (B) Luciferase assays using the vectors encoding putative conserved target sites of FSCN1 3′-UTR identified with the TargetScan database: four conserved sites for miR-145 and one site for miR-133a. (C) Luciferase assays using the mutated vectors in which the specific sites targeted by the microRNAs were deleted.
Effect of miR-145 and miR-133a transfection on cell growth, invasion, and migration activity in BC cell lines
The expression levels of miR-145 and miR-133a were extremely low in the BC cell lines compared with normal bladder epithelia (N1 and N2) (Figure 3A), suggesting that endogenous miR-145 or miR-133a in these cell lines does not affect cell viabilities. Therefore, we performed gain-of-function studies using the microRNA transfectants to investigate the functional role of miR-145 and miR-133a. The XTT cell-growth assay showed significant cell-growth inhibitions in miR-145 and miR133a transfectant compared with the controls from BOY and T24 cell lines (BOY, 86.6±1.6, 65.7±0.3, 100±0.6, respectively, P<0.0001; and T24, 87.4±0.6, 69.5±1.5, 100±0.9, respectively, P<0.0005; Figure 3B). The wound-healing assay showed significant cell migration inhibitions in miR-145 and miR133a transfectant (BOY, 59.0±3.5, 58.1±3.4, 100.0±2.4, respectively, P<0.0001; and T24, 74.5±2.5, 72.3±4.0, 100.0±2.7, respectively, P<0.0001; Figure 3C). The matrigel invasion assay also showed significant cell invasion inhibitions in the transfectants compared with the control from the BOY cell lines (35.625±4.606, 24.000±4.516, 182.000±18.678, P<0.0001; Figure 3D). However, no significant difference was observed in the miR-145 and miR-133a transfectants from T24 cell lines (173.875±16.607, 140.125±6.799, 167.000±27.367; Figure 3D). We did not subject KK47 cell line to these experiments because it showed a focal growth and it was not suitable for the experiments.
(A) miR-145 and miR-133a expression in BC cell lines (BOY, T24, KK47) and normal human bladder mucosa (N1 and N2). (B–D) Effect of cell viabilities in miR-145 and miR-133a transfectants: (B) cell growth determined by the XTT assay; (C) cell migration activity determined by the wound-healing assay; and (D) cell invasion activity determined by the matrigel invasion assay in BOY and T24 cell lines transfected with miR-145 and miR-133a. *P<0.005, **P<0.0001.
Effect of FSCN1 knockdown on cell growth, invasion, and migration activity in BC cell lines
The expression levels of FSCN1 mRNA were more than three-fold higher in BC cell lines than in the control (normal human bladder RNA). To examine the functional role of FSCN1, we performed loss of function studies using si-FSCN1-transfected T24 cell lines, which showed higher FSCN1 mRNA expression levels compared with BOY (Figure 4A, upper). We did not subject KK47 cell line to these experiments because it showed a focal growth and it was not suitable for the experiments. FSCN1 protein expression was repressed by si-FSCN1 transfection (Figure 4A, lower). The XTT assay revealed significant cell-growth inhibition in the three si-FSCN1 transfectants in comparison with that in the si-control transfectant (% of cell viability; 69.9±1.3, 88.7±2.0, 58.0±1.4, and 100.0±1.3, respectively, P<0.0001; Figure 4B). The wound-healing assay also showed significant cell migration inhibitions in the si-FSCN1 transfectant compared with the counterparts (% of wound closure; 70.9±2.5, 49.4±2.5, 34.2±2.6, and 100.0±2.6, respectively, P<0.0001; Figure 4C). The matrigel invasion assay showed that the number of invading cell was significantly decreased in the si-FSCN1 transfectant compared with the counterparts (% of cell invasion; 39.0±4.6, 35.1±2.9, 18.3±2.5, and 100.0±3.9, respectively, P<0.0001; Figure 4D).
FSCN1-knockdown effect on BC cell viability by si-RNA: (A) upper, FSCN1 mRNA expression in three BC cell lines (BOY, T24, KK47) by real-time RT–PCR; (A) lower, western blot revealed that FSCN1 protein was markedly decreased in three si-FSCN1 transfectants compared with the controls; (B) cell growth as revealed by the XTT assay; (C) cell migration activity by the wound-healing assay; and (D) cell invasion activity by the matrigel invasion assay in T24 cell lines transfected with si-FSCN1. si-FSCN1-transfected T24 cell lines exhibited a significant decrease in cell growth, migration, and invasion in comparison with the si-control transfectants. **P<0.0001.
Immunohistochemistry of FSCN1 and in situ hybridisation of miR-145 in clinical BC samples
To visualise FSCN1 expression and the related microRNA expression in a tumour lesion and surrounding normal tissues, we performed immunohistochemistry of FSCN1 and in situ hybridisation of miR-145 in FFPE tissues (Figure 5). H&E staining showed a high-grade tumour lesion surrounded by smooth muscle layers (Figure 5A). Immunohistochemistry revealed that FSCN1 was markedly expressed in the tumour lesion, whereas no expression was observed in adjacent tissues including the smooth muscle layers (Figure 5B). In contrast, miR-145 was faintly expressed in the tumour lesion with the strong expression in the smooth muscle layers (Figure 5C). The scramble-control probe showed no significant staining in either the tumour or the smooth muscle layers (Figure 5D). Figure 5E shows immunostaining of FSCN1 in a non-invasive BC (pTa) and an invasive BC with involvement of the muscularis (pT2). There was faint staining in the non-invasive BC, whereas there was strong staining of cytosol and nuclei in the invasive BC. The staining score of the invasive BC (⩾pT1) was significantly higher than that of the non-invasive BC (pTa) (1.62±0.05 vs 1.33±0.07, P=0.0055). We found no correlation between FSCN1 expression and clinico-pathological parameters except for tumour stage.
In situ hybridisation of miR-145 and immunohistochemistry examination of FSCN1 in clinical BC specimens: (A) H&E staining, tumour, and surrounding smooth muscle; (B) immunohistochemical staining of FSCN1 showing strong expression in tumour lesion; (C) in situ hybridisation of miR-145 showing faint expression in tumour lesion and strong expression in smooth muscle layer; (D) no staining by scramble-control probe; and (E) FSCN1 protein expression in invasive and non-invasive BC specimens. Low-grade bladder cancer without invasion (pTa) (upper panel, original magnification × 400). High-grade bladder cancer with involvement of the muscularis (pT2) (lower panel, original magnification × 400).
Discussion
Earlier studies showed that miR-145 and miR-133a are commonly down-regulated in several human cancers and that their transfection reduces cell proliferation of each cancer cell line (Bandrés et al, 2006; Sempere et al, 2007; Slaby et al, 2007; Szafranska et al, 2007; Ozen et al, 2008; Schepeler et al, 2008; Wang et al, 2008, 2009a, 2009b; Wong et al, 2008a; Childs et al, 2009; Friedman et al, 2009; Liu et al, 2009; Takagi et al, 2009; Tong et al, 2009; Lin et al, 2009a). Consistent with earlier studies, we found significant cell-growth inhibitions in BC cell lines transfected with miR-145 and miR-133a precursors. These results suggest that these microRNAs may have tumour suppressive functions through regulating oncogenic genes in human malignancies. Regarding BC, miR-145 was listed in two of the three earlier studies investigating microRNA signatures in BC compared with normal control (Friedman et al, 2009; Lin et al, 2009a, 2009b). Moreover, this study is the first to show that miR-133a is a down-regulated microRNA in BC. An earlier study showed that miR-133a is abundantly expressed in muscle cells, and it may have a part in regulating proliferation and differentiation (McCarthy and Esser, 2007). Regarding the target genes, there are only three earlier studies showing that miR-145 directly binds to c-Myc (Sachdeva et al, 2009) and insulin receptor substrate-1 (Shi et al, 2007), which are associated with cell proliferation and that miR-133a/b directly binds to pyruvate kinase type M2 expression, which is a potent oncogene in solid cancers (Wong et al, 2008b). Down-regulation of these microRNAs may have a critical function in BC development. Our cell invasion assay showed that there were significant decreases of invading cell number in the miR-145 and miR133a transfectants from BOY, but not from T24 BC cell lines. These results suggest that another pathway might be more crucial than FSCN1 for invasiveness in some BCs. To find the target genes, web-based software was used in the earlier studies. However, the many candidate microRNAs identified by the web-based software often make it more difficult for researchers to find the crucial target genes. In this study, we used an oligo-microarray to screen the candidates from gene expression profiles in miR-145 transfectant and found a new target gene, FSCN1, which was subsequently validated by the luciferase reporter assay. Thus, gene expression profiles from specific microRNA transfectant may be a good strategy for finding candidate genes targeted by microRNA.
FSCN1 functions in two major forms of actin-based structures: cortical cell protrusions that mediate interactions between cells and the extra-cellular matrix (ECM), cell-to-cell interactions, and cell migration; and cytoplasmic microfilamentous bundles that contribute to cell architecture and intracellular movements (Kureishy et al, 2002). The fascin–actin interaction is affected by extra-cellular cues, and certain ECM components induce bundling of actin by FSCN1 (Hashimoto et al, 2005). It is plausible that the activation of fascin through ECM substrates contributes to tumour growth, migration, and invasion. In BC, FSCN1 over-expression has been noted in three different immunohistochemistry studies (Tong et al, 2005; Karasavvidou et al, 2008; Soukup et al, 2008). Our immnohistochemical study consistently showed that the expression levels of FSCN1 were correlated with advanced tumour stage. In addition, tumour viability was markedly decreased in FSCN1-knockdown BC cell lines. These results strongly suggest that this molecule may function as an oncogene. It may be deeply associated with BC invasiveness and might be a useful staging biomarker for clinical BC.
Regarding FSCN1 regulation, several studies have reported that the actin-binding activity of fascin is inhibited by phosphorylation of residue Ser-39 by protein kinase Cα (Adams et al, 1999); β-catenin/T cell factor signalling transactivates the FSCN1 promoter in human colon carcinoma cell lines (Vignjevic et al, 2007); and FSCN1 down-regulation is associated with a decrease in β-catenin and c-erbB-2 expression (Xie et al, 2005). However, to our knowledge, there has been no earlier study reporting the interaction between FSCN1 expression and particular microRNAs. We earlier reported that miR-145 and miR-133a expressions are significantly down-regulated in BC tissue compared with normal bladder epithelium (Ichimi et al, 2009). In this study, we showed that miR-145 and miR-133a directly target FSCN1, resulting in decreased FSCN1 mRNA and its protein levels both in vitro and in clinical specimens. The question of how FSCN1 becomes over-expressed is still open, but one possible mechanism is through regulation by microRNAs. Loss of miR-145 and miR-133a, both of which are endogenous FSCN1 inhibitors, may promote aberrant expression of FSCN1 contributing to pathogenesis and progression of BC.
In summary, through our microRNA profiling in BC, we have found that FSCN1 might have an oncogenic function in BC and miR-145 and miR-133a might function as tumour suppressors through direct repression of FSCN1 in BC. As viral vector-mediated microRNA transduction might be applicable in vivo (Yang et al, 2006), our findings raise the possibility that miR-145 and miR-133a may have potential therapeutic value in BC patients. In addition, FSCN1 may be a potential target for gene therapy of BC. As down-regulation of miR-145 and miR-133a and over-expression of FSCN1 were commonly identified in various human malignancies, our findings may be crucial events in the development throughout human malignancies.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adams JC, Clelland JD, Collett GD, Matsumura F, Yamashiro S, Zhang L (1999) Cell–matrix adhesions differentially regulate fascin phosphorylation. Mol Biol Cell 10: 4177–4190
Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J (2006) Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 5: 29
Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF (2009) Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 174: 736–745
Darnel AD, Behmoaram E, Vollmer RT, Corcos J, Bijian K, Sircar K, Su J, Jiao J, Alaoui-Jamali MA, Bismar TA (2009) Fascin regulates prostate cancer cell invasion and is associated with metastasis and biochemical failure in prostate cancer. Clin Cancer Res 15: 1376–1383
Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA (2009) The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res 69: 2623–2629
Goncharuk VN, Ross JS, Carlson JA (2002) Actin-binding protein fascin expression in skin neoplasia. J Cutan Pathol 29: 430–438
Grothey A, Hashizume R, Ji H, Tubb BE, Patrick Jr CW, Yu D, Mooney EE, McCrea PD (2000) C-erbB-2/HER-2 upregulates fascin, an actin-bundling protein associated with cell motility, in human breast cancer cell lines. Oncogene 19: 4864–4875
Hashimoto Y, Skacel M, Adams JC (2005) Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int J Biochem Cell Biol 37: 1787–1804
Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N (2009) Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 125: 345–352
Jawhari AU, Buda A, Jenkins M, Shehzad K, Sarraf C, Noda M, Farthing MJ, Pignatelli M, Adams JC (2003) Fascin, an actin-bundling protein, modulates colonic epithelial cell invasiveness and differentiation in vitro. Am J Pathol 162: 69–80
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71–96
Karasavvidou F, Barbanis S, Pappa D, Moutzouris G, Tzortzis V, Melekos MD, Koukoulis G (2008) Fascin determination in urothelial carcinomas of the urinary bladder: a marker of invasiveness. Arch Pathol Lab Med 132: 1912–1915
Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH (2006) In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 3: 27–29
Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC (2002) Fascins, and their roles in cell structure and function. Bioessays 24: 350–361
Kwak C, Ku JH, Park JY, Lee E, Lee SE, Lee C (2004) Initial tumor stage and grade as a predictive factor for recurrence in patients with stage T1 grade 3 bladder cancer. J Urol 171: 149–152
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858
Lin CK, Chao TK, Yu CP, Yu MH, Jin JS (2009b) The expression of six biomarkers in the four most common ovarian cancers: correlation with clinicopathological parameters. APMIS 117: 162–175
Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, Huang L (2009a) MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol 181: 1372–1380
Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li H, DiRenzo J, Korc M, Cole CN, Bak M, Kauppinen S, Dmitrovsky E (2009) Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res 15: 1177–1183
Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH (2003) Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol 16: 902–912
McCarthy JJ, Esser KA (2007) MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 102: 306–313
Ozen M, Creighton CJ, Ozdemir M, Ittmann M (2008) Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27: 1788–1793
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108
Pelosi G, Pastorino U, Pasini F, Maissoneuve P, Fraggetta F, Iannucci A, Sonzogni A, De Manzoni G, Terzi A, Durante E, Bresaola E, Pezzella F, Viale G (2003) Independent prognostic value of fascin immunoreactivity in stage I non-small cell lung cancer. Br J Cancer 88: 537–547
Qiu D, Katanoda K, Marugame T, Sobue T (2009) A joinpoint regression analysis of long-term trends in cancer mortality in Japan (1958–2004). Int J Cancer 124: 443–448
Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava EM, Kosma VM (1999) Reduced expression of á catenin is associated with poor prognosis in colorectal carcinoma. J Clin Pathol 52: 10–16
Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY (2009) p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 106: 3207–3212
Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, Kauppinen S, Ørntoft TF, Andersen CL (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68: 6416–6424
Schickel R, Boyerinas B, Park SM, Peter ME (2008) MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27: 5959–5974
Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN (2007) Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67: 11612–11620
Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R (2007) Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem 282: 32582–32590
Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R (2007) Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72: 397–402
Sobin LH, Wittekind C (2002) TNM Classification of Malignant Tumours, 6th edn. International Union Against Cancer (UICC). New York: Wiley-Liss. 199–202
Soukup V, Babjuk M, Dusková J, Pesl M, Szakáczová M, Zámecník L (2008) Does the expression of fascin-1 and tumor subclassification help to assess the risk of recurrence and progression in t1 urothelial urinary bladder carcinoma? Urol Int 80: 413–418
Sugimoto T, Seki N, Shimizu S, Kikkawa N, Tsukada J, Shimada H, Sasaki K, Hanazawa T, Okamoto Y, Hata A (2009) The galanin signaling cascade is a candidate pathway regulating oncogenesis in human squamous cell carcinoma. Genes Chromosomes Cancer 48: 132–142
Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA (2007) MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26: 4442–4452
Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y (2009) Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 77: 12–21
Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J (2009) MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther 16: 206–216
Tong GX, Yee H, Chiriboga L, Hernandez O, Waisman J (2005) Fascin-1 expression in papillary and invasive urothelial carcinomas of the urinary bladder. Hum Pathol 36: 741–766
van Rhijn BW, van der Poel HG, van der Kwast TH (2005) Urine markers for bladder cancer surveillance: a systematic review. Eur Urol 47: 736–748
Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875
Vignjevic D, Schoumacher M, Gavert N, Janssen KP, Jih G, Laé M, Louvard D, Ben-Ze'ev A, Robine S (2007) Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res 67: 6844–6853
Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF (2009b) Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers 26: 27–34
Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, Zhou H, Zhao RC (2009a) miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol 34: 1461–1466
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM (2008) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3: e2557
Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing Yuen A, Wai-Man Ng R, Ignace Wei W (2008a) Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J Cancer 123: 251–257
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI (2008b) Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14: 2588–2592
Xie JJ, Xu LY, Zhang HH, Cai WJ, Mai RQ, Xie YM, Yang ZM, Niu YD, Shen ZY, Li EM (2005) Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem Biophys Res Commun 337: 355–362
Yang L, Bailey L, Baltimore D, Wang P (2006) Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci USA 103: 11479–11484
Zhang H, Xu L, Xiao D, Xie J, Zeng H, Cai W, Niu Y, Yang Z, Shen Z, Li E (2006) Fascin is a potential biomarker for early-stage oesophageal squamous cell carcinoma. J Clin Pathol 59: 958–964
Acknowledgements
We thank Ms M Miyazaki for her excellent laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Chiyomaru, T., Enokida, H., Tatarano, S. et al. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer 102, 883–891 (2010). https://doi.org/10.1038/sj.bjc.6605570
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605570
Keywords
This article is cited by
-
Effects of hypoxia and reoxygenation on mitochondrial functions and transcriptional profiles of isolated brain and muscle porcine cells
Scientific Reports (2022)
-
Genetic variants in miR-145 gene are associated with the risk of asthma in Taiwan
Scientific Reports (2022)
-
FSCN1 induced PTPRF-dependent tumor microenvironment inflammatory reprogramming promotes lung adenocarcinoma progression via regulating macrophagic glycolysis
Cellular Oncology (2022)
-
CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway
Cell Death & Disease (2021)
-
Targeting the cytoskeleton against metastatic dissemination
Cancer and Metastasis Reviews (2021)