Abstract
Changes in circulating miRNA profiles have been associated with different diseases. Here we demonstrate the circulating miRNA profile in serum of HCV infected individuals using a microRNA array that profiles the expression of 940 miRNAs. Serum samples from two HCV genotype - 1 and two HCV genotype - 3 infected individuals were compared with healthy controls. Expression levels of miR-134, miR-198, miR-320c and miR-483-5p that were commonly upregulated in case of both genotypes were validated in 36 individual patient serum samples. Serum miR-134, miR-320c and miR-483-5p were significantly upregulated during HCV infection. miR-320c and miR-483-5p were also upregulated in HCV- JFH1 infected cells and cell culture supernatant. Pathway analysis of putative target genes of these miRNAs indicated involvement of PI3K-Akt, NFKB and MAPK signaling pathways. Results revealed novel insights on the role of circulating miRNAs in mediating pathogenesis in HCV-infected cells.
Similar content being viewed by others
Introduction
Hepatitis C virus1 was discovered in 1989 as a major cause of chronic non-A non-B hepatitis2. It is a positive strand RNA virus classified in the Hepacivirus within the Flaviviridae family. The single stranded uncapped RNA is 9.6 kb in size and codes for a single polyprotein of 3011 amino acid residues. 130–200 million people are estimated to be infected worldwide with highest prevalence in North Africa and South Asia. Most people fail to clear HCV infection and are at risk of developing hepatic fibrosis, cirrhosis and hepatocellular carcinoma3. Chronic hepatitis is one of the leading causes of liver cirrhosis and hepatocellular carcinoma. The currently recommended therapy is a 24 or 48 week regime of pegylated interferon alpha along with ribavirin. However sustained virological response is seen in only 42%–80% patients, depending on the viral genotype4 and the treatment is associated with undesirable side effects.
Recent findings have implicated miRNAs in various aspects of HCV infection and hepatocellular carcinoma. miRNAs are 22 nt long regulatory RNAs that assemble into RISC complexes, bind target mRNAs and induce mRNA degradation or suppress protein translation. Alterations in miRNA expression profiles upon viral infection may either favour or inhibit different stages of the viral life cycle. In case of HCV, there is no evidence of viral-encoded miRNAs. However the cellular miRNA, miR-122 is a well known regulator of HCV translation and replication and is also being pursued as an antiviral5,6. While miR-122 binds to HCV RNA and favours HCV translation and replication, some miRNAs like miR-199a* and those modulated by interferon beta (IFN-β) target the HCV genome and inhibit HCV RNA production7,8. Thus they play an important role in IFN-mediated antiviral response. HCV infection has also been found to induce miR-155 expression that promotes hepatocyte proliferation and tumorogenesis by activating Wnt signaling, an example of miRNA-mediated pathogenesis9. A recent microarray analysis of human hepatoma cells infected with HCV identified 108 miRNAs that were differentially expressed in response to HCV infection10. It was demonstrated that when synthetic mimics or inhibitors of some of these miRNAs were used, the HCV RNA abundance in JFH1 infected cells was affected.
miRNAs are found not just within cells but also in serum and other body fluids. The function of these extracellular circulating miRNAs is not well understood. They may be released due to tissue damage as in the case of miR-122 and miR-192, which are indicative of liver degeneration in mice in response to drug treatment11. They may be mediators of cell-cell communication as seen in Epstein Barr virus infected cells where miRNAs secreted by infected cells are taken up by monocyte-derived dendritic cells repressing certain target genes in the dendritic cells12. Changes in circulating miRNA profiles have been associated with different diseases. They may be used as potential biomarkers because of their stability in body fluids, sensitive detection by quantitative PCR and non-invasiveness in obtaining samples of body fluids13. For example, serum miR-21, miR-122 and miR-223 were seen to be elevated in patients with hepatocellular carcinoma or chronic hepatitis and these miRNAs could be used as diagnostic biomarkers for liver injury14. Profiling of circulating miRNAs may also be used to further study any association with the mechanism of pathogenesis, tissue damage or cell-cell communication. In the present study we have examined the serum miRNA profiles of patients infected with HCV genotype 1 or genotype 3, as compared to healthy individuals.
Results
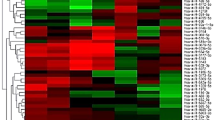
In order to identify differentially expressed miRNAs in the serum of HCV infected individuals, a miRNA microarray was carried out using serum samples from healthy controls as well as HCV infected individuals. 200 μl of serum from each of the samples was used for RNA isolation. The RNA was quantified and 500 ng of RNA each, from two healthy controls, two HCV genotype 1 infected individuals and two HCV genotype 3 infected individuals were used for hybridization on the microarray. Data obtained from microarray analysis revealed a number of miRNAs that were differentially regulated in the serum of HCV infected samples as compared to healthy controls. A representative heat map of a subset of miRNAs that were commonly upregulated in both genotype 1 and genotype 3 infected samples is shown in Figure 1. The complete list of all differentially regulated miRNAs can be found as Supplementary Table S1 online.
Four of the miRNAs that were upregulated in both genotype 1 and genotype 3 infected samples and whose expression was also reported to be modulated in HCV infected liver tissues were selected for validation by quantitative RT-PCR. These include miR-134, miR-198, miR-320c and miR-483-5p. The expression levels of these miRNAs were tested in 36 HCV infected patient serum samples and 15 healthy controls (Table 1). The average Ct values of three of these miRNAs, miR-134, miR-320c and miR-483-5p were significantly upregulated (~2 Cts, p-value < 0.05) in HCV infected samples as compared to controls (Figure 2). miR-320c showed the maximum and most significant upregulation (~2.5 Cts, p-value = 0.0008) followed by miR-134 (~1.8 Cts, p-value = 0.0137) and miR-483-5p (~1 Cts, p-value = 0.0439).
Serum miRNA expression in HCV infected and control serum samples.
The expression levels of mir-134, miR-198, miR-320c and miR-483-5p were profiled in the serum of 36 HCV infected patient samples and 15 controls by qRT-PCR assay for validation of the microarray data. The relative expression of miRNAs is expressed as the difference in threshold cycle number (Ct) between the samples and the controls. A single unit difference in Ct between the samples and controls (ΔCt) represents a 2(ΔCt) absolute difference in the miRNA levels. * denotes a p-value < 0.05 while *** denotes a p-value < 0.001. p-values for miR-320c, miR483-5p and miR-134 are 0.0008, 0.0439 and 0.0137, respectively.
Serum miRNAs that are differentially regulated in HCV infection would most probably originate from the liver. Hence it was of interest to look at the levels of these miRNAs in HCV infected liver cells. For this study, RNA was isolated from JFH1 infected Huh7.5 cells followed by qRT-PCR to check for the expression levels of miR-134, miR-198, miR-320c and miR-483-5p. miR-320c and miR-483-5p were found to be significantly upregulated both within the infected cells as well as the culture supernatant of infected cells (Figure 3).
To gain some insight into the possible role of these miRNAs in liver cells, their mRNA targets were predicted and filtered for those that were predicted by at least 2 different target prediction programs. KEGG pathway analysis of putative targets of these two miRNAs as well as previously validated targets was carried out (Table 2). It indicated that some of the target genes of miR-320c and miR-483-5p were involved in PI3K-Akt signaling pathway, NFKB signaling pathway and MAPK signaling pathway. A protein-protein interaction network for these targets was established by importing the target genes into STRING and presented using Cytoscape. The top ranking nodes in the network which link different pathways were identified by Cyto-hubba and include BRCA1, SRC, NCOA1, NFKB1, ITGAL, IGF1, ELK1, NRIP1, ESR1 and EP300 (Figure 4).
Discussion
Alterations in cellular miRNA profiles in response to HCV infection have been previously reported. These miRNAs may be positive regulators of viral infection, mediators of virus induced pathogenesis or mediators of host defense against infection. Apart from intracellular miRNAs, recent studies have also demonstrated the presence of miRNAs extracellularly in various body fluids. Serum miRNAs have been found to be stable and readily detectable by real time PCR, which have made them attractive candidates for diagnosis, prognosis and targets for therapy in different diseases. The origin and function of these circulating miRNAs remains to be explored. Our interest was to study the circulating miRNAs in HCV infection and correlate it with mechanisms of virus-induced pathogenesis.
From literature, it is known that some specific miRNAs, such as miR-122, miR-155, miR-125b, miR-16 and miR-34a, are differentially regulated in the serum of HCV infected patients15,16,17. Incidentally, a very recent report showed the serum levels of miR-92a and miR-20a as markers of HCV mediated liver disease progression using a serum-miRNA specific array18. However, our study profiles the expression of 940 human miRNAs to identify all deregulated miRNAs in the infected serum and provide a broader perspective to the analysis.
A number of miRNAs were found to be differentially regulated in HCV infected patient samples as compared to healthy controls. We analysed the expression levels of four miRNAs, miR-134, miR-198, miR-320c and miR-483-5p by qPCR. Among them, miR-134, miR-320c and miR-483-5p were found to be significantly upregulated in the serum of individuals with HCV infection. miR-320c and miR-483-5p have been previously detected in the serum of patients with liver cirrhosis19, whether they are specific to HCV induced infection is yet to be studied. Interestingly, from literature, it has been observed that hsa-miR-134 is repressed in HCV-infected liver tissue samples20,21, but it was upregulated in the HCV infected serum samples used in our study. Such reciprocal changes between miRNAs in serum and liver tissue have been observed before in drug induced liver injury11. However, there are also reports of miRNAs that are upregulated in the serum as well as infected or cancerous tissue22. Thus changes in liver tissue miRNA profile upon HCV infection can be reflected in the serum in either way.
We were interested in studying the intracellular levels of the serum miRNAs validated in our study during HCV infection. When the expression levels of miR-134, miR-198, miR-320c and miR-483-5p in HCV infected cells were studied, miR-320c and miR-483-5p were found to be significantly upregulated not only within the infected cells but also in the cell culture supernatant. The putative target genes of miR-320c and miR-483-5p were obtained using miRecords.
Predicted targets of miR-320c included ATM, ICAM1, ESR1 and NR3C1, which are involved in the NFKB signaling pathway; GHR, INSR, ITGB3 classified in the PI3K-Akt signaling pathway; and CACAN1A classified in the MAPK signaling pathway. These pathways are well known to be targeted by HCV in chronic liver disease leading to hepatoceullar carcinoma23. ADAMTS5, a regulator of cell invasion is also a known target of miR-320c24. In case of miR-483-5p, validated targets included SRF25, a modulator of VEGF signaling and ERK126, a member of the Ras-ERK intracellular signaling pathway that is modulated by HCV proteins such as NS5A27. The association of these target genes within the cellular pathway network was visualized using Cytoscape and the most crucial genes in the network which link all the pathways together were identified using Cyto-hubba. Hubba analysis indicated that the regulators of apoptosis BRCA1, ESR1, IGF1, NFKB1 and SRC play an important role in the regulatory events mediated by these miRNAs. Thus upregulation of miR-320c and miR-483-5p could be a mechanism for regulation of multiple pathways that either contribute to HCV pathogenesis or play a role in host defense against infection. Since they are also secreted into the cell culture supernatant, one could speculate their involvement in cell-cell communication or as indicators of HCV-induced liver damage.
Thus our study underlines the importance of serum miRNA in understanding disease pathogenesis. Their presence in the serum could be either due to tissue damage or an inherent cellular mechanism for transporting specific miRNAs for cell-cell communication. They might also be used as predictive biomarkers that reflect the pathophysiology of the body during HCV infection. Within infected cells, they could regulate various pathways playing a role in HCV-mediated pathogenesis.
Methods
Use of human serum
All experiments with HCV infected patient serum were performed in accordance with relevant guidelines and regulations, following the approval of the licensing committee of the Yashoda Hospital, Secunderabad, India.
Sample processing and total RNA isolation
Samples were collected from Yashoda Hospital, Somajiguda, Hyderabad, India after informed consent from the subjects. Whole blood, collected into falcon tubes, was allowed to stand overnight at 4°C. Separation of serum was accomplished by centrifugation at 3000 rpm for 10 min at 4°C. The serum obtained was transferred into new tubes, followed by a second centrifugation at 3000 rpm for 10 min at 4°C. Total RNA was isolated from 200 μl serum using miRNeasy mini kit according to the manufacturer's protocol (Qiagen). The RNA was quantified by NanoDrop 2000c Spectrophotometer. Serum from two HCV genotype 1 infected patients (one male, one female, age group: 40–70 years), two HCV genotype 3 infected patients (one male, one female, age: 40–70 years) and two healthy subjects (one male, one female, age: 40–70 years) were used for the microarray.
RNA quality control
Total RNA integrity was assessed using RNA 6000 Nano Lab Chip on the 2100 Bioanalyzer (Agilent, Palo Alto, CA) following the manufacturer's protocol. Total RNA purity was assessed by the NanoDrop® ND-1000 UV-Vis Spectrophotometer (Nanodrop technologies, Rockland, USA). We considered RNA to be good quality based on the rRNA 28S/18S ratio and RNA integrity number (RIN).
Labeling and microarray hybridization
Microarray experiments were carried out at Genotypic Technology Pvt. Ltd, Bangalore, India. The miRNA labeling was performed using miRNA Complete Labeling and Hyb Kit (Agilent Technologies, Part Number: 5190-0456). The total RNA sample was diluted to 50 ng/ul in nuclease free water. About 100 ng of total RNA was dephosphorylated using Calf Intestinal Alkaline Phosphatase (CIP) master mix (Agilent Technologies, Part Number: 5190-0456) by incubating at 37°C for 30 minutes. The dephosphorylated miRNA sample was denatured by adding dimethyl sulfoxide and heating at 100°C for 10 minutes and transferred to ice-water bath. The ligation master mix (Agilent Technologies, Part Number: 5190-0456) containing Cyanine 3-pCp was added to the denatured miRNA sample and incubated at 16°C for 2 hours. The Cyanine 3-pCp labeled miRNA sample was dried completely in the vacuum concentrator (Eppendorf, Concentrator Plus, Catalog Number 5305) at 45°C for 2 hours. The dried sample was resuspended in 18 μl of nuclease free water and mixed with Hybridization Mix containing blocking solution (Agilent Technologies, Part Number: 5190-0456) and Hi-RPM Hybridization Buffer (Agilent Technologies, Part Number: 5190-0456) and incubated at 100°C for 5 minutes followed by snap chill on ice for 5 minutes. The samples were hybridized on the Human miRNA 8 × 15 k Arrays AMADID: 021827 and the hybridization was carried out at 55°C for 20 hours. After hybridization, the slides were washed using Gene Expression Wash Buffer1 (Agilent Technologies, Part number: 5188-5325) at room temperature for 5 minutes and Gene Expression Wash Buffer 2 (Agilent Technologies, Part Number: 5188-5326) at 37°C for 5 minutes. The slides were then washed with acetonitrile for 30 seconds. The microarray slide was scanned using Agilent Scanner (Agilent Technologies, Part Number: G2565CA).
Microarray data analysis
Data extraction from images was done using Feature Extraction software v 10.7 of Agilent. Feature extracted data was analyzed using GeneSpring GX version 11.5 software from Agilent. Normalization of the data was done in GeneSpring GX using the 90th percentile shift and normalization to specific samples. [Percentile shift normalization is a global normalization, where the locations of all the spot intensities in an array are adjusted. This normalization takes each column in an experiment independently and computes the percentile of the expression values for this array, across all spots (where n has a range from 0–100 and n = 90 is the median)]. It subtracts this value from the expression value of each entity. Significant differentially expressed miRNA up- and downregulated within the group of samples were identified.
HCV infection of Huh7.5 cells
Huh7.5 monolayer cells (90% confluent) were transfected with JFH1 RNA28 using Lipofectamine 2000 reagent (Invitrogen) in antibiotic free medium. Five hours post-transfection, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum was added. The cells and cell culture supernatant were harvested after 48 hours using TRI reagent (Sigma) and Tri reagent BD (Sigma) respectively for total RNA isolation and qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR)
The TaqMan microRNA Assay System was used for miRNA detection and quantification. Briefly, the RT reaction was performed in a final volume of 7.5 μl containing 0.75 μl of 10× RT buffer, 2.5 μl of total RNA (30 ng), 1.5 μl of 5× miRNA-specific RT primer, 0.075 μl of 100 mM dNTP, 0.1 μl of 20U/μl RNase inhibitor and 0.5 μl of MultiScribe reverse transcriptase (50 U/μl). The reaction condition was 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. Real-time PCR was then performed in a total reaction volume of 10 μl containing 0.66 μl of RT product, 5 μl of 10× TaqMan Universal PCR master mix and 0.5 μl of the primer and probe mix from the TaqMan MicroRNA Assay Kit. The reaction condition was 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Statistical significance was determined by using the two-tailed Mann–Whitney non-parametric test.
miRNA target prediction and Pathway analysis
miRNA targets were predicted using miRecords (http://mirecords.biolead.org/) and the pathways in which these genes lie were identified using KEGG database29. Protein-protein interaction networks for miRNA target genes were retrieved from STRING30, a database of known and predicted protein interactions. Networks were presented using Cytoscape31 and the hubs of networks were identified using a plug in of Cytoscape called cyto-Hubba.
References
Lee, M. H. et al. Community and personal risk factors for hepatitis C virus infection: a survey of 23,820 residents in Taiwan in 1991–2. Gut 60, 688–694 (2011).
Kuo, G. et al. An assay for circulating antibodies to a major etiologic virus of human non-A., non-B hepatitis. Science 244, 362–364 (1989).
McGivern, D. R. & Lemon, S. M. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene 30, 1969–1983 (2011).
Poynard, T. et al. A comparison of fibrosis progression in chronic liver diseases. J Hepatol 38, 257–265 (2003).
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581 (2005).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Murakami, Y., Aly, H. H., Tajima, A., Inoue, I. & Shimotohno, K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol 50, 453–460 (2009).
Pedersen, I. M. et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449, 919–922 (2007).
Zhang, Y. et al. Hepatitis C Virus-induced upregulation of miR-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology (2012).
Lee, M. H. et al. Hepatitis C virus infection and increased risk of cerebrovascular disease. Stroke; a journal of cerebral circulation 41, 2894–2900 (2010).
Wang, K. et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 106, 4402–4407 (2009).
Pegtel, D. M. et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107, 6328–6333 (2010).
Etheridge, A., Lee, I., Hood, L., Galas, D. & Wang, K. Extracellular microRNA: a new source of biomarkers. Mutat Res 717, 85–90 (2011).
Xu, J. et al. Circulating microRNAs, miR-21, miR-122 and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Molecular carcinogenesis 50, 136–142 (2011).
Bihrer, V. et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol 106, 1663–1669 (2011).
Bihrer, V. et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One 6, e26971 (2011).
Cermelli, S., Ruggieri, A., Marrero, J. A., Ioannou, G. N. & Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One 6, e23937 (2011).
Shrivastava, S. et al. Upregulation of circulating miR-20a is correlated with hepatitis C virus mediated liver disease progression. Hepatology (2013).
Li, L. M. et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 70, 9798–9807 (2010).
Varnholt, H. et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 47, 1223–1232 (2008).
Ura, S. et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49, 1098–1112 (2009).
Wang, L. G. & Gu, J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol 36, e61–67 (2012).
Arzumanyan, A., Reis, H. M. & Feitelson, M. A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 13, 123–135 (2012).
Ukai, T., Sato, M., Akutsu, H., Umezawa, A. & Mochida, J. MicroRNA-199a-3p, microRNA-193b and microRNA-320c are correlated to aging and regulate human cartilage metabolism. J Orthop Res 30, 1915–1922 (2012).
Qiao, Y. et al. MiR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett 585, 3095–3100 (2011).
Wang, L. et al. MiR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett 586, 1312–1317 (2012).
Macdonald, A. et al. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J Biol Chem 278, 17775–17784 (2003).
Wakita, T. et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11, 791–796 (2005).
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. & Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40, D109–114 (2012).
Szklarczyk, D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39, D561–568 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Acknowledgements
We thank Dr Takaji Wakita, Dr Charles M Rice and Dr Ralf Bartenschlager for the cell lines and plasmid constructs. We thank our lab members and Sharath Pattar for their helpful discussion. This work was supported by grant from Department of Biotechnology, (DBT), Government of India. SS is supported by the research fellowship from Council of Scientific and Industrial Research (CSIR) and GK is supported by the postdoctoral fellowship of DBT, India.
Author information
Authors and Affiliations
Contributions
S.S., S.D. wrote the main manuscript text and S.S., K.G. prepared figures 1–4, M.C., B.R., M.N.K. determined the genotypes and virus titer. S.S., K.G. performed experiments. S.S. and S.D. designed and analyzed the experiment and results.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Shwetha, S., Gouthamchandra, K., Chandra, M. et al. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep 3, 1555 (2013). https://doi.org/10.1038/srep01555
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01555
This article is cited by
-
SQSTM1/p62 promotes miR-198 loading into extracellular vesicles and its autophagy-related secretion
Human Cell (2022)
-
Factors influencing circulating microRNAs as biomarkers for liver diseases
Molecular Biology Reports (2022)
-
Differentially expressed plasmatic microRNAs in Brazilian patients with Coronavirus disease 2019 (COVID-19): preliminary results
Molecular Biology Reports (2022)
-
Inhibition of mitochondrial complex II in neuronal cells triggers unique pathways culminating in autophagy with implications for neurodegeneration
Scientific Reports (2021)
-
Expression profiling of miRNA-196a biomarker in naïve hepatitis C virus-infected and Sofosbuvir plus Daclatasvir-treated patients
Archives of Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.