Abstract
Bicomponent pore-forming leukocidins are a family of potent toxins secreted by Staphylococcus aureus, which target white blood cells preferentially and consist of an S- and an F-component. The S-component recognizes a receptor on the host cell, enabling high-affinity binding to the cell surface, after which the toxins form a pore that penetrates the cell lipid bilayer. Until now, six different leukocidins have been described, some of which are host and cell specific. Here, we identify and characterise a novel S. aureus leukocidin; LukPQ. LukPQ is encoded on a 45 kb prophage (ΦSaeq1) found in six different clonal lineages, almost exclusively in strains cultured from equids. We show that LukPQ is a potent and specific killer of equine neutrophils and identify equine-CXCRA and CXCR2 as its target receptors. Although the S-component (LukP) is highly similar to the S-component of LukED, the species specificity of LukPQ and LukED differs. By forming non-canonical toxin pairs, we identify that the F-component contributes to the observed host tropism of LukPQ, thereby challenging the current paradigm that leukocidin specificity is driven solely by the S-component.
Similar content being viewed by others
Introduction
The human and animal pathogen Staphylococcus aureus is capable of colonizing and infecting a broad range of host species. S. aureus has been shown to adapt to its hosts through acquisition of mobile genetic elements and the introduction of allelic variation through chromosomal mutations. For example, ruminant and equine S. aureus strains have acquired pathogenicity islands encoding host-specific variants of von Willebrand factor-binding protein1,2 and recently a single nucleotide polymorphism in the dltB gene was shown to make a human S. aureus strain capable of infecting rabbits3.
Leukocidins are a family of bicomponent pore-forming toxins contributing to S. aureus pathogenicity. Currently there are six known leukocidins of S. aureus (HlgAB, HlgCB, LukAB/HG, LukED, Panton-Valentine leukocidin (LukSF-PV/PVL), and LukMF’), all consisting of two subunits (an S- and an F-component) that together induce pore formation. In the current model of pore formation, the S-component first binds to a specific receptor on the cell surface, after which the F-component can associate to form octameric beta-barrel pores in the host cell membrane4. Both gamma-hemolysins (hlgAB and hlgCB) and lukAB/HG are encoded in the core genome of S. aureus, while lukED is located on a common pathogenicity island (vSaβ). In contrast, pvl and lukMF’ are located on prophages4. While pvl is mostly found in S. aureus isolates from human origin, lukMF’ is almost exclusively harboured by strains from ruminant origin5,6,7,8. Corresponding with their distribution, these leukocidins display specific host tropisms, explained by the high-affinity interaction of the toxins with receptor molecules which differ between host species9,10,11,12. This leads to large differences in leukocidin activity between host species. For example, PVL has been shown to lyse neutrophils from rabbits and humans, but to have no effect on Java monkey neutrophils13, while LukMF´ is highly toxic to ruminant neutrophils, but not to human neutrophils14,15.
Here, we describe a novel phage-encoded member of the S. aureus bicomponent leukocidin family named LukPQ, which shares 91% and 80% amino-acid sequence identity with LukE and LukD respectively. We show that LukPQ is strongly associated with S. aureus strains isolated from Equidae (horses and donkeys) and, in accordance with this distribution, preferentially kills neutrophils from equine origin with a higher efficiency than its closest relative LukED. We identify the equine-CXCRA and CXCR2 as receptors for the S-component, but, in contrast to the current paradigm, we show that the observed host specificity is not solely determined by the S-component, but also in part by the F-component.
Results
LukPQ: a new phage encoded leukocidin associated with equids
In the genome sequences of a collection of S. aureus clonal complex (CC)133 from horses and donkeys we identified a 45 kb prophage (named: ΦSaeq1) that displayed considerable sequence similarity and synteny to the previously reported phage ΦSaov3, which encodes the ruminant LukMF’ (Fig. 1a). ΦSaeq1 was highly conserved among equid CC133 strains and was integrated at a position ~0.5 Mb into the chromosome at approximately the same site as ΦSaov1 and SaPIbov1 in ED133 and RF122, respectively2. ΦSaeq1 encoded a novel leukocidin, close to the amidase genes of the phage (Fig. 1a). As the strains carrying this new variant were isolated from two species of Equidae, we propose that the new toxin be named LukPQ (P for Paardachtigen, Dutch for Equidae) and use isolate 3711 as a reference strain for describing this phage and leukocidin locus. Phylogenetic analysis of LukPQ in comparison to the rest of the leukocidin family showed that LukP was most closely related to LukE (91% amino acid identity), whereas LukQ was most similar to the ruminant associated LukF’ (83% amino acid identity), but also shared 80% amino acid sequence with LukD (Fig. 1b). Molecular modelling of LukP and LukQ confirmed that both subunits adopt classical leukocidin folds (Supplementary Fig. 1). To further validate the association with equids we screened our collection of sequenced genomes by BLASTn and found lukPQ with 99–100% nucleotide identity in 29 isolates from 5 different clonal complexes (CC1, CC133, CC350, CC522, CC1660), and from a broad geographical distribution of countries (Brazil, Switzerland, Tunisia, United Kingdom), primarily from equid hosts, but also in 5 isolates from ruminants (Supplementary Table 1). In the majority of positive isolates (96%), lukPQ was present on a phage, but in two strains from Brazilian buffalo, lukPQ was flanked by only two phage-related genes (amidase and holin); the remainder of the phage was not present in the genome of these strains. Between CCs, the phage encoding lukPQ showed considerable variation, but lukPQ was highly conserved, showing only few SNPs, which were associated with clonal lineage (Supplementary Table 1), comparable to what has been shown for pvl-encoding phages16.
(a) Comparison of the novel prophage ΦSaeq1 in isolate 3711, carrying the equid specific lukPQ, with ΦSaov3 (ruminant strain ED133) and ΦSa2 (human PVL strain MW2). Areas of red show regions conserved between the sequences and homologous coding DNA sequences are marked in the same colour. (b) Phylogenetic tree based on amino acid sequences of mature leukocidins, with alpha-hemolysin as an outgroup.
We estimated the prevalence of lukPQ in an international collection of equid S. aureus isolates (The Netherlands (unpublished), Austria17, the United States18, Sweden19, Portugal20, Italy21 and Spain22) using a PCR assay to identify the three prophage-encoded leukocidins (lukSF-PV, lukMF’ and lukPQ). lukPQ was present in 29 out of 194 strains (15%, 95% CI: 10 to 21%) from the Netherlands, Italy and Portugal, whereas lukSF-PV and lukMF’ were only found once and twice, respectively (Supplementary Table 2). Between isolate collections, the prevalence of lukPQ differed considerably. In the Dutch collection LukPQ was found in 25 of 74 isolates (34%); interestingly this included 11 out of 21 isolates (52%) from the spa-type t011 (CC398) - a lineage that has been reported to be specifically associated with horses23.
LukPQ preferentially kills horse neutrophils
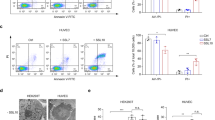
As there was evidence for the association of LukPQ with equid hosts, we sought to identify if it exhibited specific activity against horse neutrophils, leukocytes known to be instrumental in the host defence against S. aureus24. Equine, bovine and human neutrophils were incubated with the three prophage-encoded leukocidins with an assumed host specificity (LukPQ (putatively equid), LukMF’ (ruminant) and LukSF-PV (human)) and pore formation was quantified in a dose dependent manner. Equine neutrophils were highly susceptible to LukPQ-induced lysis with a half-maximal lytic concentration (EC50) of 0.46 nM (±SD 0.23) (Fig. 2a). This was higher than the EC50 of LukMF’ on bovine neutrophils (0.08 nM (±SD 0.02), p < 0.001) (Fig. 2b)14, but significantly lower than the EC50 of LukSF-PV on human neutrophils (1.63 nM (±SD 0.66), p = 0.006) (Fig. 2c). Both LukMF’ and LukSF-PV were unable to induce pore formation in equine neutrophils, emphasizing their described host restrictions13,15. LukPQ, however, was able to permeabilise both human and bovine neutrophils, but at significantly higher EC50’s (45.82 nM (±SD 11.10) and 5.68 nM (±SD 1.64) respectively, both p < 0.0001) (Fig. 2a).
LukPQ acts on CXCRA and CXCR2
Based on the high degree of similarity (91% amino-acid identity) between receptor binding components LukE and LukP, we hypothesized that the most likely receptors for LukPQ would comprise CXCR1, CXCR2, CCR5, and the Duffy antigen receptor (DARC) analogous to LukED10,11. We cloned and expressed the equine homologues of these receptors (the putative CXCR1 homologue in equids is CXCRA25) and CCR2 and C5aR in HEK293T cells and exposed these cells to LukPQ and LukED. This identified CXCRA and CXCR2 as the major receptors for LukPQ with EC50’s of 5.81 nM (±SD 3.9) and 3.46 nM (±SD 1.09) respectively (Fig. 3a). Pore formation through CCR5 was less efficient (EC50 > 270 nM), while no pore formation was observed in HEK293T cells expressing DARC, C5aR or CCR2. Additionally, we showed that LukED also permeabilises cells carrying equine CXCRA, CXCR2 and CCR5 at efficiencies similar to LukPQ (EC50’s of 8.97 nM (±SD 5.74) for CXCRA and 6.93 nM (±SD 3.24) for CXCR2) (Fig. 3b). To further investigate the interaction of the S-component LukP with the CXCRA and CXCR2 receptors, we tested its ability to functionally antagonize stimulation of these receptors. The horse CXCRA and CXCR2 receptors expressed on HEK293T cells were shown to respond to stimulation with human CXCL6 and CXCL8 (ligands for CXCRA) or CXCL5 and CXCL6 (ligands for CXCR2) by intracellular calcium mobilization (Fig. 3c). After priming the CXCRA and CXCR2 transfected cells with LukP, we observed that intracellular calcium mobilization upon stimulation with their specific cytokines was significantly reduced. This suggests that LukP interacts with CXCRA and CXCR2 at the ligand-binding site of these receptors and has immunomodulatory properties when present as a single component. Alternatively, it may be that LukP induces internalization of the receptor, resulting in less surface receptor and therefore in reduced calcium mobilization.
(a) Pore formation in HEK293T cells stably transfected with equine CCR2, CCR5, C5aR, CXCRA, CXCR2 and the Duffy antigen receptor (DARC) upon treatment with LukPQ. Mean percentages of permeable cells ± SD are shown (n = 3). (b) HEK293T cells stably transfected with equine CXCRA, CXCR2 and CCR5 were incubated with LukED and analysed for pore formation. Mean percentages of permeable cells ± SD are shown (n = 3). (c) Relative calcium mobilization by CXCRA and CXCR2 transfected HEK293T cells preincubated with buffer or 10 μg/ml LukP upon stimulation with CXCL5, CXCL6 and CXCL8. Bars indicate SD with n = 3. Statistically significant effects of preincubation with LukP are indicated. **P < 0.01 and *P < 0.05. Pre-incubation with LukP resulted in a significant decrease in calcium mobilization in both CXCRA and CXCR2 cells stimulated with CXCL6 (p < 0.01 and p < 0.05 respectively), and in CXCR2 cells stimulated with CXCL5 (p < 0.05). A trend was seen in CXCRA cells stimulated with CXCL8 (p = 0.06).
LukPQ and LukED exhibit different species specificities
While the presence of lukPQ was enriched in equid isolates, the closely related lukED, located on a pathogenicity island, is present in most S. aureus isolates5. We identified that all of the sequenced equid strains in our collection (Supplementary Table 1) that harboured lukPQ also harboured lukED, although in CC133 strains, the lukE gene was disrupted by a nonsense mutation in amino acid position 174, as has been reported for other CC133 strains2. In order to assess the additional value of LukPQ in equid isolates in comparison to the ubiquitously present LukED, we compared the cytotoxicity of both toxins on equine, bovine and human neutrophils. Interestingly, when comparing EC50 values, LukED is a significantly less potent killer of equine neutrophils than LukPQ with an EC50 of 6.62 nM (±SD 4.45) (p = 0.004) (Fig. 4a). This finding was not apparent in the data from the receptors expressed in HEK293T cells, where LukPQ and LukED displayed almost equal toxicity (p = 0.73 for CXCRA and p = 0.46 for CXCR2 expressing cells) (Fig. 3). Human neutrophils are permeabilised significantly more efficiently by LukED than by LukPQ (p < 0.001), while for bovine neutrophils the increased efficiency of LukED is minimal and non-significant (p = 0.079) (Fig. 4b and c).
The F-component is involved in host-specificity
Next, because of the high degree of similarity between LukP and LukE, we analysed the effects of the non-canonical toxin pairs LukPD and LukEQ on the different neutrophils. LukEQ showed a significant increase in pore formation in equine neutrophils as compared to LukED with an EC50 of 0.74 nM (±SD 0.59) (p = 0.007) and was as potent as the native pair LukPQ (p = 0.98) (Fig. 4a). This suggests that LukQ is involved in host specificity to horse neutrophils, a finding that was corroborated by the poor activity of other non-canonical pair: LukPD. Against bovine neutrophils, LukEQ and LukED displayed equal activity with EC50’s of 1.51 nM (±SD 0.47) and 2.17 nM (±SD 1.31) respectively (p = 0.9), and the EC50 of LukEQ was marginally better than the EC50 of LukPQ (5.68 nM (±SD 1.64) p = 0.032, Fig. 4b), suggesting that LukE has a greater specificity for bovine neutrophils than LukP. Finally, against human neutrophils the canonical combination LukED displayed significantly higher activity than all other pairs (p < 0.001), which displayed low-level activity– suggesting that the targeting of human neutrophils requires both LukE and LukD (Fig. 4C). Taken together, the results demonstrate that both the S and F-components of LukED and LukPQ are involved in host specificity and importantly, reveal a previously unrecognised role for a leukocidin F-component in host specificity.
Discussion
In this study, we describe a new member of the S. aureus bicomponent pore-forming toxin family: LukPQ, which is phage-encoded and associated with equid hosts. In accordance with its host distribution, we showed that LukPQ displays an enhanced cytotoxicity towards equine neutrophils. This suggests an important role for LukPQ in the evasion of the host defence mechanism of S. aureus in equids, in line with the assumed function of other phage-encoded leukocidins (LukMF’ and PVL) that have a similarly host-specific function13,14 and distribution26 (Supplementary Table 3). S. aureus regularly causes problems in equine hospitals, leading primarily to joint, skin and wound infections27. Patient-to-patient transmission and outbreaks within equine hospitals as well as zoonotic transmission have been documented18,19,28,29,30. Recently, an epidemic subclone of CC398 MRSA was shown to have spread within and between equine hospitals23. This subclone consisted almost exclusively of spa-type t011 strains, which in our study had a high prevalence of LukPQ. Leukocidins protect S. aureus from migrating neutrophils, which are the hosts first line of defence24, by creating a protective zone around it14, enabling it to reproduce after initial entry into a new host. Likewise, LukPQ may enhance the transmission between equid hosts, driving the success of this clone in equine hospitals. However, further evaluation of the clinical impact of LukPQ in equid infection is required.
The γ-hemolysins and LukED target a broad host range and are widely distributed amongst S. aureus lineages15,31,32,33, consistent with a more generalist function. LukPQ demonstrates host specificity, but has a broader host range than LukMF’ and PVL as at higher concentrations it is capable of lysing bovine and to some extent human neutrophils. We demonstrated that LukPQ targets CXCRA and CXCR2, the equine CXCL8 (IL-8) receptors expressed on neutrophils34, as well as CCR5, albeit with lower affinity. While the receptor tropism of LukPQ and LukED is similar, we found a species-dependent difference in cytotoxicity towards neutrophils: LukPQ is more toxic to equine neutrophils than LukED, while the opposite is true for human neutrophils. The S-components LukE and LukP are highly similar. The rim domain, particularly the DR4 region, of the S-component of the toxin is important for receptor binding. Consistent with their shared receptor specificity, the DR4 regions of LukE and LukP are almost identical, whilst that of LukM, which binds CCR1, is considerably different (Fig. 5A). The DR4 region of HlgA, which also binds CXCR1 and CXCR2, is highly similar to that of LukE and LukP, but lacks a GS insertion which may explain why HlgA also targets CCR2 rather than CCR512 (Fig. 5A).
(a) Structure-guided alignment of the DR4 region (highlighted yellow) in the rim domain of LukE, LukP, HlgA and LukM. (b) Homology model of the LukPQ heterodimer with LukP as a cartoon and LukQ as a surface representation. Residues unique to LukQ, but identical between LukD and LukF’ are coloured yellow; residues that differ in all three toxins are coloured cyan. The position of isoleucine 285 in the rim domain is annotated.
Analysis of the effect of the non-canonical pairs LukPD and LukEQ suggests that the F-components LukD and LukQ (which share only 80% sequence identity) are the key determinants of the difference in activity between LukPQ and LukED against equine and human neutrophils, whereas LukD and LukQ have equal specificity for bovine neutrophils. Comparing LukQ, LukD and LukF’ identifies 20 residues that are unique to LukQ, but which are conserved between LukD and LukF’ and a further 13 residues that differ between all three toxins (Supplementary Fig. 2). Some of these LukQ-unique residues are found in the likely interface for binding with LukP, and one of the unique residues, I285 in the LukQ rim domain, maps to a position previously identified in LukF-PV as important for interaction with the cell membrane35 (Fig. 5B). Further studies involving chimeric F-components may yield insight in the actual importance of these residues. Still, the variable residues do not group onto one specific surface, so it is unclear whether the host specificity mediated through LukD and LukQ stems from the interaction between the F-component and the S-component, or from the interaction between the F-component and the cell membrane. Although F-components do not interact with the cognate GPCR receptors of the leukocidins9, LukF-PV has been suggested to bind to an F-component receptor prior to complex formation, possibly explaining the differences in activity between canonical and non-canonical combinations of F-components with LukS-PV36,37. In the case of LukPQ, we found no significant binding of the F-component to equine neutrophils (Supplementary Fig. 3), suggesting that interaction with an F-component receptor prior to pore-formation is unlikely. However, there is a possibility that F-components recruit a different receptor to the complex of alternating S- and F-components during the pore formation process. Involvement of such a receptor might explain the difference in species specificity of LukED and LukPQ. Future studies will be needed to elucidate the molecular mechanism of pore formation and identify all players involved in the process38.
In conclusion, we describe a novel leukocidin with a high sequence similarity to LukED, but we show that the small differences in amino-acid sequence of the S-components in combination with a different F-component leads to a substantial change in affinity for neutrophils of various host species, and therefore to host specificity.
Methods
Ethics statement
All experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all human blood donors in accordance with the Declaration of Helsinki. The medical ethics committee of the University Medical Center Utrecht (The Netherlands) approved the use of human venous blood for this study. The use of blood from cattle was approved by the Ethical Committee for Animal Experiments of the Utrecht University (Permit No. DEC2012.II.10.152) and conducted according to national regulations.
Bacterial strains and genome sequencing
Strains used in this study were isolated in the course of previous and on-going studies39,40 or collected as part of routine surveillance. Genomic DNA was extracted with the MasterPure Gram-positive DNA purification kit (Cambio, United Kingdom). HiSeq sequencing was performed according to the manufacturer’s protocol (Illumina, Inc., United States). Phage identification was performed using PHAST41. The nucleotide sequence of the LukPQ positive phage from strain 3711 has been deposited in the Sequence Read Archive database in the European Nucleotide Archive (LT671578).
To estimate the prevalence of the three phage encoded leukocidins, previously reported collections of horse isolates17,18,20,21,22,42 and a selection of isolates from an undescribed Dutch collection were screened by PCR (see Supplementary Methods).
Leukocyte isolation
Bovine blood was collected from the coccygeal vein of healthy Holstein Friesian cows using a sterile blood collection system with EDTA anti-coagulant (BD Vacutainer). Neutrophils were isolated by using Percoll (1.09176 g/l) centrifugation14. Human blood was collected in heparin tubes from healthy volunteers and neutrophils were isolated by Ficoll/Histopaque centrifugation43. Blood was collected from healthy horses during the slaughter process (and immediately upon death) in tubes containing 3 mM EDTA anticoagulant. Equine neutrophils were isolated using 70 and 85% Percoll gradients as described44.
Cloning, expression and purification of recombinant proteins
Recombinant LukP, LukQ, and LukD proteins were generated in E. coli according to methods described previously45. See Supplementary Methods for details and primer sequences. Recombinant PVL and LukMF’ used in this study were generated as reported previously9,14. Recombinant LukE was kindly provided by Thomas Henry (Lyon, France)46.
Cloning of receptor expressing plasmids
Horse genomic DNA was obtained from Zyagen (San Diego, USA). Equine chemokine receptors CXCRA, CXCR2, CCR2, CCR5, C5aR1, and the predicted Duffy antigen receptor (DARC) were amplified from equine genomic DNA by PCR using PfuTurbo DNA polymerase (Stratagene). Primers and accession numbers are listed in Supplementary Table 6. Exons encoding DARC were assembled using overlap extension PCR. All coding sequences were cloned into the pIRESpuro3 vector (Clontech) according to methods described elsewhere12. The human Ga16 cDNA (pCISG16 plasmid) was kindly provided by Melvin I. Simon47. The Ga16 gene was recloned in between the BstBI and EcoRV sites of the pIREShyg3 vector (Clontech) using the following primers:
5′-AACTATTTCGAAGCCGCCACCATGGCCCGCTCGCTGACCTG-3′ and
5′-ATCGAGGATATCTCACAGCAGGTTGATCTCGTC-3′.
Cell lines and Transfections
HEK293T cells (a human embryonic kidney cell line obtained from the American Type Culture Collection) were maintained in DMEM supplemented with 10% FCS and 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293T cells were stably transfected with human Gα16 plasmids prior to transfection with equine receptor encoding plasmids. Cells were selected for Gα16 expression using 250 μg/ml hygromycin. Transfections with pIRESpuro3 and pIREShyg3-Gα16 plasmids were performed as described12.
Cell permeability assays
HEK293T cells and neutrophils (3 × 106 cells/ml) were incubated with recombinant LukPQ, PVL, LukED, or LukMF’14 in a volume of 50 μl in RPMI, containing 0.05% human serum albumin (Sanquin) for 30 minutes at 37 °C, 5% CO2. Cells were analyzed by flow cytometry and pore formation was defined as intracellular staining by 4′,6-diamidino-2-phenylindole (DAPI). Equimolar concentrations of S- and F-components were applied in all assays. For analysis, the percentage of DAPI-positive cells incubated with buffer (spontaneously permeabilised cells) was subtracted from the percentage of DAPI-positive cells that were incubated with toxin. Half maximal lytic concentrations (EC50) were calculated using nonlinear regression analyses in Prism6 (Graphpad Software Inc., USA). EC50 data were log transformed and analysed using one-way ANOVA, followed by Tukey’s multiple comparison test.
Intracellular Calcium mobilization assays
Calcium mobilization assays with CXCRA and CXCR2 HEK293T cells were performed as described48, with slight modifications. Cells were resuspended to 5 × 106 cells/ml in Hanks’ Balanced Salt Solution (HBSS) supplemented with 10 mM HEPES, 0.05% HSA and 25 μM Probenecid and were loaded with 2 μM Fluo-3-AM (Invitrogen) for 1 hour at 37 °C while shaking. Cells were washed, resuspended to 5 × 106 cells/ml in the described HBSS buffer and incubated with buffer or 10 μg/mL LukP for 30 minutes at room temperature. Cells were stimulated with different concentrations of CXCL5, CXCL6, and CXCL8. The increase in calcium mobilization was assessed by flow cytometry for 10 seconds before and up to 70 seconds after addition of the stimulus. Relative calcium mobilization was calculated by dividing the mean fluorescence after stimulation by that of the background. The effect of stimulation with or without LukP was assessed using a general linear model, modelling the interaction between concentration of the ligand and presence or absence of LukP on the relative calcium mobilization.
Computational analysis and leukocidin homology modelling
Homology models were generated with Modeller (v9.14)49. For LukM, LukF’, LukP and LukQ templates were derived either from the Homstraad database or from LukE (PDB ID: 3ROH50); whilst human CXCR1 (PDB ID: 2LNL51) was used as a template for equine CXCRA. Models were created using thorough MD optimisation and very thorough VTFM optimisation before analysis with the integral DOPE function of Modeller. The model of highest initial quality was further refined and improved using SCWRL451,52 and Molprobity53. Structural alignments were performed using the superposition function of PYMOL (Schrodinger Inc.). All structural images were generated with PYMOL. Sequence alignments and pairwise identities were determined with Clustal Omega54. Topology predictions for the membrane spanning receptor proteins were calculated using the Constrained Consensus Topology prediction server55,56.
Additional Information
How to cite this article: Koop, G. et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci. Rep. 7, 40660; doi: 10.1038/srep40660 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Viana, D. et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77, 1583–1594 (2010).
Guinane, C. M. et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2, 454–466 (2010).
Viana, D. et al. A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat. Genet. 47, 361–366 (2015).
Alonzo, F. III . & Torres, V. J. The bicomponent pore-forming leucocidins of Staphylococcus aureus . Microbiol. Mol. Biol. Rev. 78, 199–230 (2014).
McCarthy, A. J. & Lindsay, J. A. Staphylococcus aureus innate immune evasion is lineage-specific: A bioinfomatics study. Infect. Genet. Evol. 19, 7–14 (2013).
Yamada, T. et al. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 110, 97–103 (2005).
Herron-Olson, L., Fitzgerald, J. R., Musser, J. M. & Kapur, V. Molecular correlates of host specialization in Staphylococcus aureus . PLoS ONE 2 (2007).
Schlotter, K. et al. Leukocidin genes lukF-P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet. Res. 43 (2012).
Spaan, A. N. et al. The staphylococcal toxin Panton-Valentine Leukocidin targets human C5a receptors. Cell Host Microbe 13, 584–594 (2013).
Alonzo, F. III et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55 (2013).
Reyes-Robles, T. et al. Staphylococcus aureus Leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to Kill leukocytes and promote infection. Cell Host Microbe 14, 453–459 (2013).
Spaan, A. N. et al. The staphylococcal toxins γ-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat. Commun. 5, 5438 (2014).
Löffler, B. et al. Staphylococcus aureus Panton-Valentine Leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6 (2010).
Vrieling, M. et al. Bovine Staphylococcus aureus secretes the leukocidin LukMF’ to kill migrating neutrophils through CCR1. mBio 6 (2015).
Barrio, M. B., Rainard, P. & Prévost, G. LukM/LukF′-PV is the most active Staphylococcus aureus leukotoxin on bovine neutrophils. Microbes Infect. 8, 2068–2074 (2006).
Boakes, E. et al. Distinct bacteriophages encoding panton-valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J. Clin. Microbiol. 49, 684–692 (2011).
Loncaric, I. et al. Identification and characterization of methicillin-resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Vet. Microbiol. 168, 381–387 (2014).
Van Balen, J. et al. Molecular epidemiology of environmental MRSA at an equine teaching hospital: Introduction, circulation and maintenance. Vet. Res. 45 (2014).
Bergström, K., Aspan, A., Landén, A., Johnston, C. & Grönlund-Andersson, U. The first nosocomial outbreak of methicillin-resistant Staphylococcus aureus in horses in Sweden. Acta Vet. Scand. 54 (2012).
Couto, N. et al. Biocide and antimicrobial susceptibility of methicillin-resistant Staphylococcal isolates from horses. Vet. Microbiol. 166, 299–303 (2013).
Mallardo, K., Nizza, S., Fiorito, F., Pagnini, U. & De Martino, L. A comparative evaluation of methicillin-resistant staphylococci isolated from harness racing-horses, breeding mares and riding-horses in Italy. Asian Pac. J. Trop. Biomed. 3, 169–173 (2013).
Gómez-Sanz, E. et al. First detection of methicillin-resistant Staphylococcus aureus ST398 and Staphylococcus pseudintermedius ST68 from hospitalized equines in Spain. Zoonoses Public Health 61, 192–201 (2014).
Abdelbary, M. M. H. et al. Phylogenetic analysis of Staphylococcus aureus CC398 reveals a sub-lineage epidemiologically associated with infections in horses. PLoS ONE 9 (2014).
Rigby, K. M. & DeLeo, F. R. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin. Immun. 34, 237–259 (2012).
Widdison, S. et al. The bovine chemokine receptors and their mRNA abundance in mononuclear phagocytes. BMC Genomics 11 (2010).
Koymans, K. J., Vrieling, M., Gorham, R. D. & van Strijp, J. A. G. In Current Topics in Microbiology and Immunology (ed Compans, R. W. et al.) 1–49 (Springer Berlin Heidelberg, Berlin, Heidelberg, 2016).
Weese, J. S. & van Duijkeren, E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet. Microbiol. 140, 418–429 (2010).
van Duijkeren, E. et al. Methicillin-resistant Staphylococcus aureus in horses and horse personnel: An investigation of several outbreaks. Vet. Microbiol. 141, 96–102 (2010).
Schwaber, M. J. et al. Clonal transmission of a rare methicillin-resistant Staphylococcus aureus genotype between horses and staff at a veterinary teaching hospital. Vet. Microbiol. 162, 907–911 (2013).
Sieber, S. et al. Evolution of multidrug-resistant Staphylococcus aureus infections in horses and colonized personnel in an equine clinic between 2005 and 2010. Microb. Drug Resist. 17, 471–478 (2011).
Siwicki, A. K. et al. In vitro effect of staphylococcal leukocidins (LukE, LukD) on the proliferative responses of blood lymphocytes in dog (Canis familiaris). Bull. Vet. Inst. Pulawy 47, 395–401 (2003).
Bownik, A. In vitro effects of staphylococcal leukocidin LukE/LukD on the proliferative ability of lymphocytes isolated from common carp (Cyprinus carpio L.). Fish Shellfish Immunol. 20, 656–659 (2006).
Spaan, A. N. et al. Differential interaction of the staphylococcal toxins Panton-Valentine Leukocidin and γ-hemolysin CB with human C5a receptors. J. Immunol. 195, 1034–1043 (2015).
Brooks, A. C., Rickards, K. J. & Cunningham, F. M. CXCL8 attenuates chemoattractant-induced equine neutrophil migration. Vet. Immunol. Immunopathol. 139, 141–147 (2011).
Monma, N., Nguyen, V. T., Kaneko, J., Higuchi, H. & Kamio, Y. Essential residues, W177 and R198, of LukF for phosphatidylcholine-binding and pore-formation by staphylococcal γ-hemolysin on human erythrocyte membranes. J. Biochem. 136, 427–431 (2004).
Meyer, F., Girardot, R., Piémont, Y., Prévost, G. & Colin, D. A. Analysis of the specificity of panton-valentine leucocidin and gamma-hemolysin F component binding. Infect. Immun. 77, 266–273 (2009).
Prevost, G. et al. Panton-valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect. Immun. 63, 4121–4129 (1995).
Yamashita, D. et al. Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins. Nat. Commun. 5 (2014).
Gharsa, H. et al. High diversity of genetic lineages and virulence genes in nasal Staphylococcus aureus isolates from donkeys destined to food consumption in Tunisia with predominance of the ruminant associated CC133 lineage. BMC Vet. Res. 8 (2012).
Aires-de-Sousa, M. et al. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microbiol. 73, 3845–3849 (2007).
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J. & Wishart, D. S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 39, W347–W352 (2011).
Bergström, K., Bengtsson, B., Nyman, A. & Grönlund Andersson, U. Longitudinal study of horses for carriage of methicillin-resistant Staphylococcus aureus following wound infections. Vet. Microbiol. 163, 388–391 (2013).
Surewaard, B. G., van Strijp, J. A. & Nijland, R. Studying interactions of Staphylococcus aureus with neutrophils by flow cytometry and time lapse microscopy. J. Vis. Exp. 77 (2013).
Siemsen, D. W., Schepetkin, I. A., Kirpotina, L. N., Lei, B. & Quinn, M. T. Neutrophil isolation from nonhuman species. Methods Mol. Biol. 412, 21–34 (2007).
Ko, Y. P. et al. Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog. 9, 1–13 (2013).
Perret, M. et al. Cross-talk between Staphylococcus aureus leukocidins-intoxicated macrophages and lung epithelial cells triggers chemokine secretion in an inflammasome-dependent manner. Cell. Microbiol. 14, 1019–1036 (2012).
Amatruda, T. T. III., Steele, D. A., Slepak, V. Z. & Simon, M. I. Ga16, a G protein a subunit specifically expressed in hematopoietic cells. Proc. Natl. Acad. Sci. USA 88, 5587–5591 (1991).
De Haas, C. J. C. et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199, 687–695 (2004).
Shi, J., Blundell, T. L. & Mizuguchi, K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257 (2001).
Nocadello, S. et al. Crystal structures of the components of the Staphylococcus aureus leukotoxin ED. Acta Crystallograph. Sect. D, Struct. Biol. 72, 113–120 (2015).
Park, S. H. et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491, 779–783 (2012).
Krivov, G. G., Shapovalov, M. V. & Dunbrack, R. L. Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins Struct. Funct. Bioinform. 77, 778–795 (2009).
Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallograph. Sect. D Biolog. Cryst. 66, 12–21 (2010).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7 (2011).
Dobson, L., Langó, T., Reményi, I. & Tusnády, G. E. Expediting topology data gathering for the TOPDB database. Nucleic Acids Res. 43, D283–D289 (2015).
Dobson, L., Reményi, I. & Tusnády, G. E. The human transmembrane proteome. Biol. Direct 10 (2015).
Acknowledgements
This work was supported by a Medical Research Council Partnership Grant (G1001787/1) held at the University of Cambridge (MAH, RNZ). GK acknowledges the receipt of a fellowship from the OECD Co-operative Research Programme: Biological Resource Management for Sustainable Agricultural Systems in 2014. MV was supported by the ALTANT project of the Ministry of Economic Affairs of the Dutch Government. JRF was supported by a project grant (BB/K00638X/1) and institute strategic grant funding (ISP3) from the Biotechnology and Biological Sciences Research Council (UK). LSCL is a Wellcome Trust Research Training Fellow and DMLS is a Cambridge Gates Scholar; the work by LSCL, MNLS and ERC was funded in part by the NIHR Cambridge BRC. EMH’s position was funded by Health Innovation Challenge Fund, WT098600, HICF-T5-342. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust.
Author information
Authors and Affiliations
Contributions
G.K., R.Z., E.M.H. and M.A.H. designed the study. G.K. and E.M.H. discovered LukPQ as an equid specific new leukocidin and, independently, E.J.R. discovered the equid phage. M.V., D.M.L.S., L.S.C.L., E.M.H., M.A.H., E.R.C., C.H., K.K. designed the experiments. M.V., G.W., C.R., X.B., N.G., N.H., A.J.T., D.M.L.S., L.S.C.L., C.H. and K.K. performed experiments. G.K. and E.M.H. performed bioinformatics analyses. T.M. created protein models of leukocidins and receptors. G.K.P., J.A.W., J.R.F., C.T., A.S.W., A.L., I.L., A.E.H., K.B., L.D.M., H.L., C.P., H.D.L., K.B.S. and H.G. provided strains or genome sequences. H.M.K. contributed to the literature review. G.K., M.V., E.M.H., T.M., J.R.F., J.A.G.S. and M.A.H. contributed to interpretation of the data. M.V., G.K., E.M.H. and T.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Koop, G., Vrieling, M., Storisteanu, D. et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci Rep 7, 40660 (2017). https://doi.org/10.1038/srep40660
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40660
This article is cited by
-
S. pseudintermedius and S. aureus lineages with transmission ability circulate as causative agents of infections in pets for years
BMC Veterinary Research (2021)
-
Staphylococcus aureus isolates from Eurasian Beavers (Castor fiber) carry a novel phage-borne bicomponent leukocidin related to the Panton-Valentine leukocidin
Scientific Reports (2021)
-
Characterization of Staphylococcus aureus isolated from milk samples of dairy cows in small holder farms of North-Western Ethiopia
BMC Veterinary Research (2018)
-
Leukotoxin and pyrogenic toxin Superantigen gene backgrounds in bloodstream and wound Staphylococcus aureus isolates from eastern region of China
BMC Infectious Diseases (2018)
-
Population genomics of bacterial host adaptation
Nature Reviews Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.