Abstract
Background:
Male breast cancer (MBC) is still poorly understood with a large proportion arising in families with a history of breast cancer. Genomic studies have focused on germline determinants of MBC risk, with minimal knowledge of somatic changes in these cancers.
Methods:
Using a TruSeq amplicon cancer panel, this study evaluated 48 familial MBCs (3 BRCA1 germline mutant, 17 BRCA2 germline mutant and 28 BRCAX) for hotspot somatic mutations and copy number changes in 48 common cancer genes.
Results:
Twelve missense mutations included nine PIK3CA mutations (seven in BRCAX patients), two TP53 mutations (both in BRCA2 patients) and one PTEN mutation. Common gains were seen in GNAS (34.1%) and losses were seen in GNAQ (36.4%), ABL1 (47.7%) and ATM (34.1%). Gains of HRAS (37.5% vs 3%, P=0.006), STK11 (25.0% vs 0%, P=0.01) and SMARCB1 (18.8% vs 0%, P=0.04) and the loss of RB1 (43.8% vs 13%, P=0.03) were specific to BRCA2 tumours.
Conclusions:
This study is the first to perform high-throughput somatic sequencing on familial MBCs. Overall, PIK3CA mutations are most commonly seen, with fewer TP53 and PTEN mutations, similar to the profile seen in luminal A female breast cancers. Differences in mutation profiles and patterns of gene gains/losses are seen between BRCA2 (associated with TP53/PTEN mutations, loss of RB1 and gain of HRAS, STK11 and SMARCB1) and BRCAX (associated with PIK3CA mutations) tumours, suggesting that BRCA2 and BRCAX MBCs may be distinct and arise from different tumour pathways. This has implications on potential therapies, depending on the BRCA status of MBC patients.
Similar content being viewed by others
Main
Recent advances in next-generation sequencing technologies have made it possible to interrogate the molecular characteristics of individual cancers. Within breast cancer research, perhaps, the best contemporary example is the recent analysis of 466 breast cancers by The Cancer Genome Atlas Network that integrated analysis from various molecular platforms to produce a comprehensive portrait of genetic and epigenetic alterations (Cancer Genome Atlas Network, 2012). Analysis revealed convergent changes leading to common gene circuits that correlated with luminal, HER2 and basal phenotypes as defined by mRNA profiling. Furthermore and importantly, from a therapeutic standpoint, a greater knowledge of genomic and potentially targetable drivers was ascertained.
While the TCGA study contained eight male breast cancers (MBCs), the analysis neither segregated nor commented on specific alterations in males. This is reflective of much of MBC research where the study and treatment of these rarer tumours have been extrapolated from findings concluded from female breast cancer (FBC) studies. Overall, MBC cancers comprise <1% of all breast cancers but account for greater mortality (Weiss et al, 2005; Korde et al, 2010). Traditionally, these tumours are thought to be most similar to peri/postmenopausal FBC with a mean/median age at diagnosis 5–10 years later than FBC with a high proportion of invasive ductal carcinomas of no special type and high frequency of oestrogen/progesterone receptor (ER/PgR) positivity (Giordano et al, 2002; Deb et al, 2012a). However, unlike FBC, there is a lower proportion of tumours of basal and possibly HER2 phenotypes (Bloom et al, 2001; Muir et al, 2003) and an absence of early onset cancers (<40 years of age) (Deb et al, 2012a). Although a significant proportion of MBCs arise in a setting of familial breast and ovarian cancer, the effect of being a BRCA mutation carrier is different from female gene carriers with a relative high penetrance seen in BRCA2 male carriers (10.3%) but very low penetrance in BRCA1 male carriers (1.2%) (Deb et al, 2012a).
Little is known about the risk factors and biology for MBC, and to date most molecular studies have examined the germline for specific predisposing genes. There are few somatic studies that have interrogated chromosomal changes largely through array-based CGH, with some reported differences compared with FBC (Tirkkonen et al, 1999; Rudlowski et al, 2006; Johansson et al, 2011; Tommasi et al, 2011). Only seven studies have specifically investigated MBCs comprising a total of 208 males (Anelli et al, 1995; Dawson et al, 1996; Hiort et al, 1996; Kwiatkowska et al, 2002; Benvenuti et al, 2008; Cancer Genome Atlas Network, 2012; Deb et al, 2013), with all but one study not reporting on BRCA status or family history of the patients. Furthermore, only a limited panel of genes have been examined, including PIK3CA, TP53, KRAS, BRAF, androgen receptor (AR) and BRCA2 mutations with some differences again noted from FBC. While older studies suggest a similar frequency of TP53 mutations (25–41% range in MBCs) (Anelli et al, 1995; Dawson et al, 1996) and BRCA2 alterations (21%) (Kwiatkowska et al, 2002) between MBC and FBCs, other MBCs studies have been inconsistent with regard to the frequency of KRAS mutations (0% vs 12%) (Dawson et al, 1996; Deb et al, 2013). Furthermore, some mutations, such as the PIK3CA E547K mutation, appear to be overrepresented and potentially specific to MBCs (Deb et al, 2013). As an extension from our previous study, we have therefore taken advantage of new technologies that are able to parallel sequence formalin-fixed, paraffin-embedded tissue and have profiled 48 familial MBCs (28 BRCAX, 17 BRCA2 and 3 BRCA1) using a 48 gene panel that includes hotspot regions of 15 of the 20 most commonly mutated genes in FBC, including those above in addition to AKT1, ALK1, APC, ATM, CDH1, CTNBB1, NOTCH1, PTEN, RB1 and SMAD4. Although the somatic mutation landscape of MBC is relatively unknown, the panel also includes genes commonly mutated in other cancers to test against. This is the most comprehensive mutational analysis performed on familial MBC to date and aims to: (1) report the type and frequency of these mutations in MBC, (2) identify the number of driver mutations in MBC and compare these with FBC, (3) identify potential mutations specific to MBC, (4) examine copy number variation (CNV) of these gene and (5) determine the genomic relationship with MBC phenotype and assess whether there are any clinicopathologic correlates. The aim of this study is to improve our understanding of the genomic landscape and architecture of MBC and to identify potential novel targets for therapy specific to this tumour type and assess whether similar targets is present in a subset of FBC. We also aim to further define familial MBC genomically, compare familial MBC with sporadic MBC and identify potential MBC subsets.
Materials and methods
Patients
Males with breast cancers were obtained from the kConFab repository (http://www.kconfab.org) and included cases from Australia and New Zealand diagnosed between 1980 and 2009. The criteria for admission to the kConFab study has been published previously (Loughrey et al, 2008). The flow of patients through the study, according to the REMARK criteria, is listed in Supplementary Table 1. Of the 118 cases within the kConFab registry, 58 cases were excluded because of the unavailability of tissue. Of the 60 cases where tissue was available, 12 cases had poor quality DNA or insufficient tumour tissue for DNA extraction. Clinical parameters, including disease-specific mortality, were obtained from referring clinical centres, kConFab questionnaires and state death registries. Information on pedigree, mutational status and testing were available from the kConFab central registry. Histologic classification was based on the criteria set by the World Health Organisation 2012 (Cleton-Jansen et al, 1995), and all slides and pathologic records from all cases were reviewed centrally within a single institute for tumour size, tumour grade, lymphovascular and perineural invasion. Immunohistochemistry was performed centrally for ERα, PgR, basal markers (cytokeratin (CK) 5, EGFR) and HER2 silver in situ hybridisation (SISH) and scored as per scoring systems described by Harvey et al (1999) and Wolff et al (2007) as reported previously (Deb et al, 2012b) and also listed in Supplementary Table 2. While a consensus on positive CK5 and EGFR scoring is not presently defined, all tumours that were positive showed strong staining in >10% of tumour cells. Using stratification of intrinsic phenotypes based on Nielsen et al (2004), tumours were placed into luminal (ERα positive, HER2 negative, CK5 and/or EGFR negative or positive), basal (HER2 and ERα negative; CK5 and/or EGFR positive), HER2 (HER2 positive, ERα, CK5 and EGFR negative or positive) and null/negative (HER2, ERα, CK5 and EGFR negative) phenotypes. This work was carried out with approval from the Peter MacCallum Cancer Centre Ethics Committee (Project No: 11/61).
Germline BRCA1/2 testing
Mutation testing for BRCA1 and BRCA2 mutations was performed as reported previously (Loughrey et al, 2008). Once the family mutation had been identified, all pathogenic (including splice site) variants of BRCA1 and BRCA2 were genotyped by kConFab in all available family members’ DNA.
DNA extraction
Genomic DNA was extracted from formalin-fixed, paraffin embedded (FFPE) samples. A 3 μm haematoxylin- and eosin-stained slide was cut from FFPE blocks and stained to identify for tumour-enriched areas (>80% tumour content). From the relevant area on the FFPE block, at least one 2 mm punch biopsy core was taken with 85% of samples having two cores extracted. The cores were then dewaxed and hydrated through gradient alcohol. Genomic DNA was then extracted using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) following proteinase K digestion at 56 °C for 3 days.
UDG treatment
The treatment of FFPE DNA with uracil-DNA glycosylase (New England Biolabs, Ipswich, MA, USA) was performed on the MyCycler instrument (Bio-Rad, Hercules, CA, USA). This has been demonstrated to significantly reduce sequence artefact induced by formalin fixation (Do et al, 2013). One unit of UDG was added for each 20 ng of FFPE DNA with 0.5 × of UDG buffer. The treatment conditions had two incubation steps: an initial activation at 37 °C for 2 h and an inactivation of UDG enzyme at 97 °C for 10 min.
Illumina TruSeq amplicon cancer panel
The TruSeq Amplicon Cancer Panel comprises a total of 212 amplicons from 48 genes (Supplementary Table 3) and 6 amplicons from reporter sequences (RP5-1091E12.1, RP11-286H14.8, RP11-530I17.1, RP11-350N15.4, CTC-554D6.1, C11orf65) that are simultaneously amplified in a highly multiplexed and single-tube reaction. Five microlitres at a concentration of 25 ng μl−1 of each DNA sample was used for the experiment according to the manufacturer’s instructions. The MiSeq system was used for paired end sequencing using a v1 150 bp kit (Illumina, San Diego, CA, USA). Forty-eight cases were able to examine gene mutation completely and 44 cases were able to assessCNV.
Sequencing validation
Within all samples, hot spots on TP53 (exons 5–7) PIK3CA (exons 9 and 20), AKT1 (exon 1), BRAF (exon 15) and KRAS (exon 2) genes were analysed for mutation by high-resolution melting and Sanger sequencing. The PIK3CA, AKT1, BRAF and KRAS data using Sanger sequencing for these exons in these patients has been published previously (Deb et al, 2013) (Supplementary Table 4 and Supplementary Figure 1). Mutations of other cancer samples on the same runs were also validated by Sequenom MassARRAY platform (San Diego, CA, USA) (Supplementary Table 4 and Supplementary Figure 1). Three MBC samples were also run at least two times across multiple sequence runs to examine for run-specific variation.
Bioinformatics
Primer sequences prefixing the short reads were used to assign each read to an amplicon. Global alignment was then performed between the reads and the amplicon reference sequences to identify sequence variations. Positive variants (in the original biologic sample) were identified using VarScan2 (http://varscan.sourceforge.net). DNA CNV was estimated by comparing sequence read depth between the breast cancer samples and a pseudocontrol. The control was created by averaging the normalised read depth from 20 random human samples that were derived from the same protocols and location as the cancer samples. The averaging and normalisation of the control group was performed using the baseline creation workflow in CONTRA (Li et al, 2012). Log ratios between a cancer sample and the control were then computed in 50 bp windows using CONTRA. Using >600 in-house samples, we estimated the null distribution of log ratios for each gene and each exon separately, and thereby making significant calls on genes/exons that lie at the extremes of the distributions (using a P-value cutoff of 0.05; Benjamini–Hochberg adjusted). Gains and losses were defined by a two-fold increase or decrease in reads, whereas amplification was determined by a four-fold increase. Deletions were not examined separate to losses.
Comparison of groups was made using Mann–Whitney U-test for nonparametric continuous distributions and χ2 test for threshold data. Kaplan–Meier survival curves were plotted using breast cancer-related death as the end point and compared using a log-rank test. A two-tailed P-value test was used in all analyses and a P-value or <0.05 was considered statistically significant.
Hierarchical clustering
Unsupervised hierarchical cluster analysis of log 2 ratios of copy numbers for each gene was used to detect possible unique signatures. Analysis was performed using Cluster and Tree View software written by Michael Eisen (Stanford University, Stanford, CA, USA) as published previously (Eisen et al, 1998; Makretsov et al, 2004; van de Rijn and Gilks, 2004) and Elucidean metric distance was used.
Results
Mutated genes in MBC
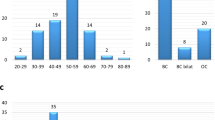
Overall, 48 tumours were sequenced with clinicopathologic variables as outlined in Table 1. A total of 11 373 mutations were identified and of these 479 were tested by an orthogonal method (Supplementary Table 4). There was a high artefactual/false-positive rate when total mutation reads were below one hundred counts or <5% of total reads for prospective germline and <125 mutation reads (and <150 read total coverage) for somatic mutations. Using this cutoff, 11 234 mutations were excluded with high sensitivity (98%) and specificity (99%) for mutation detection (Supplementary Figure 1). Overall, 98% of our amplicons had coverage of >150 reads. Subsequently, 112 variants, 15 nonsense mutations and 12 missense somatic mutations were identified.
No case had more than one somatic mutation present. The 12 mutations (Table 2 and Figure 1) in 48 cases (25%) were only present in three genes: PIK3CA (9 mutations, incidence 18.8%), TP53 (2 mutations, incidence 4.2%) and PTEN (1 mutation, incidence 2.1%). Of the nine PIK3CA mutations, seven were present in BRCAX patients (7 mutation, incidence 28–25%), with one each in BRCA1 (1 mutation, incidence 3–33%) and BRCA2 (1 mutation, incidence 17–5.9%) patients. Four PIK3CA mutations were present in exon 9 (E542K, E547K), three in exon 20 (H1047R) and two in exon 5 (N345K). The two TP53 mutations were found in exon 5 (A138P) and exon 8 (R306Q). The single PTEN mutation (E314*) was a truncating mutation in exon 8. All TP53 (2 mutation, incidence 17–11.8%) and PTEN (1 mutation, incidence 17–5.9%) mutations were found in BRCA2 cases.
All cases with mutations occurred in invasive carcinomas of no special type (IC-NST), with one PIK3CA mutation (N345K) and the single PTEN mutation occurring in IC-NSTs with a component of invasive micropapillary carcinoma (Table 2). No associations were observed between germline mutation groups and clinicopathologic and prognostic factors, including age at diagnosis, tumour size, grade, histologic subtype, hormone and HER2 receptor status, TNM stage, phenotype or disease outcome.
Allelic variants
One hundred and twelve allelic variants were identified (Table 3). There were 17 single-nucleotide polymorphisms (SNPs) present in 11 genes. Of these, the most common were the homozygous P72R (rs1042522) TP53 variant (37.5% frequency), heterozygous T1493T (rs41115) APC variant (43.8%), homozygous T1493T (rs41115) APC variant (43.8%) and the heterozygous V824V (rs2228230) PDGFRA variant (31.3%). There were no variants overrepresented in any particular BRCA subgroups and the frequency was within that reported in the general population. There was no association between variants and the previously mentioned clinicopathologic factors or cancer phenotype.
Copy number analysis
Satisfactory data were retrieved from 44 cases (3 BRCA1, 16 BRCA2 and 25 BRCAX) for copy number analysis (Figure 2A–C). Overall, out of 54 regions (48 genes and 6 reporters), the median number of genes showing copy number changes (adjusted for multiple testing) seen per sample was 9.5 (range 0–48). This did not significantly vary between BRCA1 (median 2, range 1–10, P=0.23), BRCA2 (median 10.5, range 2–36, P=0.88) and BRCAX (median 13, range 0–48, P=0.31) cases (Figure 3A). Dividing the overall cohort into three groups of low (0–4), intermediate (>4–16) and high (>16) numbers of copy number changes showed no differences in associated clinicopathologic features or disease-specific survival (Figure 3B).
(A) Number of genes gained or lost per case, stratified by BRCA status, (B) disease-specific survival stratified by volume of copy number changes per case (0–4=low, >4–16=intermediate, >16=high), (C) unsupervised cluster analysis of MBCs showing a loss predominant cluster (cluster A) and gain predominant cluster (cluster B) and ( D ) disease-specific survival of cluster A vs B.
Across the MBC cohort (Table 4), the only gain seen in >30% of cases was for GNAS (34.1%, chromosome position 20q13.3). Losses were seen in GNAQ (36.4%, 9q21), ABL1 (47.7%, 9q34.1) and ATM (34.1%, 11q22–q23), as well as the C11orf65 reporter (38.6%, 11q22.3). Analysis stratified by BRCA status (Table 4) showed differences between groups. Only three cases of BRCA1 MBCs were present with the most common changes noted being losses of ABL (67%), NOTCH1 (67%, 9q34), ATM (67%) and C11orf65 (67%). In BRCA2 cases, aside from also harbouring the common gains and losses across all MBCs, there were also gains in CTNNB1 (31.3%, 3p21), FGFR3 (31.3%, 4p16.3) and HRAS (37.5%, 11p15.5), and losses in NRAS (31.3%, 1p13.2), FBXW7 (31.3%, 4q31.3), APC (37.5%, 5q21–q22), CTC-554D6.1 reporter (37.5%, 5q22.2), RP11-286H14.8 reporter (31.3%, 7q32), PTEN (31.3%, 10q23.2), KRAS (31.3%, 12p12.1) and RB1 (43.8%, 13q14.2). In BRCAX cases, no areas of gain were seen but losses were seen in NRAS (32.0%, 1p13.2), KIT (36.0%, 4q11–q12), FIP1L1 (36.0%, 4q12), PDGFRA (36.0%, 4q12) and MET (32.0%, 7q31) on top of also the common losses seen in all MBCs. An association of BRCA2 mutation carrier status was seen with gains of HRAS (37.5% vs 3%, P=0.006), STK11 (25.0% vs 0%, P=0.01) and SMARCB1 (18.8% vs 0%, P=0.04), and the loss of RB1 (43.8% vs 13%, P=0.03). No other changes were seen specific for a BRCA subgroup.
Unsupervised clustering showed two large groups (Figure 3C). One group (group A, correlation coefficient 0.359) was defined by a predominance of gene loss, whereas the second group (group B, correlation coefficient 0.360) was defined by gene gain. The CNV differences seen in group A when compared with group B included loss of ABL1, AKT1, FGFR3, SMO, RET, FGFR1, KDR, JAK3, NOTCH1, KIT, EGFR, SMARCB1, PDGFRA, FIP1L1, SRC, FBXW7, CSF1R, STK11, FLT3, MPL, GNAQ and ALK with gain of NPM1. Comparison of the two groups showed no association with BRCA status or clinicopathologic factors including disease-specific survival (Figure 3D).
Rank comparison between copy number changes was performed within the BRCA2 (Supplementary Table 5) and BRCAX cohorts (Supplementary Table 6). Owing to the low numbers of BRCA1 cases, this group of patients was excluded. Within BRCA2 cases, the strongest correlation (r>0.9, P<0.0001) seen was between SMO (7q32.1) and SMARCB1 (22q11.23), PTPN11 (12q24.1) and CTNNB1 (3p21), CSF1R (5q32) and RET (10q11.2) and between RET and CTNNB1. In the BRCAX cohort, a correlation was seen between KDR (4q11–q12) and EGFR (7p12), ERBB4 (2q33.3–q34) and FBXW7 (4q31.3), PDGFRA/FIP1L1 (4q12) and PTEN (10q23.2) and between RB1 (13q14.2) and SMAD4 (18q21.1).
Comparison of ERBB2/HER2 SISH and copy numbers generated by MiSeq showed significant correlation (r=0.46, P<0.01) (Supplementary Figure 2). Only two instances of amplification (FGFR1 and FGFR3) were seen and not subanalysed further.
Discussion
Our data showed somatic mutations in familial MBC occur at a lower overall frequency compared with FBC (Cancer Genome Atlas Network, 2012), which is in agreement with the limited data from TCGA. However, the profile of mutations observed in this familial MBC cohort is similar to that seen in luminal/ER-positive FBCs with which they share common phenotypic features (Cancer Genome Atlas Network, 2012; Deb et al, 2012a). The most common mutations identified in MBCs (where possible to compare) are similar, albeit at lower frequencies (PIK3CA mutations (19% vs 45%) followed by TP53 (4% vs 12%) and then PTEN (2% vs 3%). Indeed, the similarities with FBC extend to the types and positions of mutations in MBC in PIK3CA with the frequencies of exon 20 mutations>exon 9 mutations>exon 5 mutations (aside from our previously reported E547K PIK3CA mutation (Deb et al, 2012a), which is rarely seen in FBC), and interestingly, the only gene mutation noted in more than one TCGA MBC sample was PIK3CA (two H1047R, one E545K). The overall rarity of TP53 mutations in our MBCs and in our analysis of the TCGA data set contrast with the historic studies by Anelli et al (1995) and Dawson et al (1996) who observed 25% (5 out of 20)–41% (12 out of 29) of MBCs harbouring TP53 mutations. Considering that these mutations are enriched in the basal and HER2 subsets of FBCs, our results are somewhat expected given that these phenotypes are more than half as frequently seen in MBCs (2% and 9%, respectively) (Deb et al, 2012a) when compared with FBC. Nevertheless, a notable difference is the absence of CDH1 mutations that are frequently reported in luminal A FBC (7%) (Corso et al, 2012). This is likely to be because of the lower incidence of lobular carcinoma in MBCs (3%) when compared with FBC (∼10%) (Cleton-Jansen et al, 1995; Deb et al, 2012a).
To date, several studies have performed array CGH analysis of MBCs or analysed oncogene amplification by multiplex ligation-dependent probe amplification. While the array CGH studies show MBCs to contain more gains than seen in FBC and more gains than losses overall (Johansson et al, 2011), our MBC cohort shows relatively equal numbers of gene gains and losses. There is some overlap in the regions noted previously, with gains at the GNAS locus (20q13.3) and losses at the ATM locus (11q22–23) also seen in MBC and FBC (Rudlowski et al, 2006; Johansson et al, 2011). Losses however of GNAQ (9q21) and ABL1 (9q34.1) have not been reported in MBC, with 9q34 loss only noted previously in ER-negative IC-NST (Loo et al, 2004), a subset rarely found in MBC.
Although an association was present between loss of ABL1 (9q34.1) and positive nodal disease (65% vs 25%, P=0.04) and between loss of GNAS and the presence of invasive papillary or micropapillary carcinoma (35% vs 6%, P=0.05), no other clinical or phenotypic association was seen either with mutations or gene CNVs. This is likely to be due to the relative homogeneity of MBCs, which are largely IC-NSTs with a luminal phenotype. Comparison with the analysis by Kornegoor et al (2012) of 110 MBCs for copy number changes in 21 genes showed some overlap with AKURA (on the same locus as GNAS) and CDH1. Unlike their study, we saw no association between FGFR1 and a younger age of onset or between ERBB2 and higher grade and mitotic count. This may be because of the cohort examined, as ours is exclusively familial MBCs that present earlier, or contain a large proportion of BRCA2 that are associated with higher mitosis and grade, whereas Kornegoor et al (2012) did not segregate cases into familial and sporadic cases or comment on the patient’s BRCA status.
Johansson et al (2011) have previously noted two subsets of MBC based on the frequency of chromosomal changes. Most MBCs fell into an MBC complex group characterised by high numbers of changes with frequent whole chromosomal arm gains/losses. These cancers grouped well with luminal complex FBCs. Although it is difficult to compare high-resolution aCGH results with copy number changes in 54 gene loci, we noted three distinct groups of cases with low (0–4), intermediate (>4–16) and high (>16) numbers of copy number gain or losses. No clinical differences were seen between these groups with relatively similar spread between BRCA2 and BRCAX cohorts. Prognostically, a very weak trend was seen showing better outcome in the low cohort compared with the intermediate and high cohorts (P=0.24). Interestingly, we also noted that genes in close chromosomal proximity shared significantly similar changes between cases consistent with the frequent whole chromosomal arm changes as seen in MBCs by Johansson et al (2011) and supporting the validity of our findings. Notably, samples that were run more than once clustered tightly, further indicating the analytical validity of the test (Supplementary Figure 3).
Several differences were observed between the BRCA subtypes. TP53 mutations, while infrequent, were restricted to tumours arising in BRCA2 carriers (11% vs 0%) with a profile more similar to luminal B cancers (Cancer Genome Atlas Network, 2012). Notably, within MBCs, BRCA2 cancers have been associated with higher grade and increased mitotic counts (Ottini et al, 2003), typical of luminal B tumours and thus may represent a novel subtype in MBCs. In contrast, BRCAX tumours had a much higher incidence of activating PIK3CA mutations (25% vs 10%), suggesting that activation of the mTOR/PIK3CA pathway may be relevant in these tumours. As most of these cancers are also ER positive and of a luminal phenotype, these features are more similar genophenotypically to the luminal A FBC s. Interestingly, in contrast to familial FBCs (Greenblatt et al, 2001), of the three BRCA1 MBCs, no TP53 mutation was seen. Whiles these numbers are low, the low penetrance of MBCs in male BRCA1 mutation carriers and a lack of tumours with basal cell phenotype suggest that the germline mutation may not be acting as a tumour driver and emphasises difference of the BRCA1 effect in MBCs compared with FBCs. Compared with familial FBCs stratified by subtypes, there was some similarities with luminal A cancers with frequent loss of 11q23 and 9q34.3. No overlap with other intrinsic subtypes was seen.
Two studies by Johansson et al (2011) and Tirkkonen et al (1999) have examined gene copy numbers in very small numbers of familial MBCs, reporting on copy number changes in three and five BRCA2 MBCs, respectively. Our BRCA2 MBCs, in comparison with other MBCs, showed novel HRAS, STK11 and SMARCB1 amplification and RB1 loss. The loss of RB1 may be because of its chromosomal proximity to the BRCA2 gene, which is supported by sporadic FBC studies showing frequent contiguous loss of RB1 and BRCA2 on the chromosome 13q12–q14 band (Cleton-Jansen et al, 1995). While accurate somatic loss of heterozygosity analysis of BRCA2 in our cases is largely restricted by availability of germline DNA, wild-type allelic loss would not be unexpected as previous studies have shown that somatic BRCA2 mutations occur frequently (21%) in sporadic MBCs (Kwiatkowska et al, 2002), and thus suggesting that BRCA2 loss is a significant driver in MBC. Interestingly, Johansson et al (2011) also noted gain of the 19p13 locus housing STK11/LKB1 in their BRCA2 MBC. As this area contains several tumour suppressor genes, it may suggest particular selection in BRCA2-deficient MBCs. Alternately, as the tumour suppressor STK11/LKB1 may also enhance ERα response, it may be that STK11/LKB1 may be oncogenic in some breast cancer subsets that may also include MBCs. Notably, in our cohort we see just as many losses of STK11/LKB1 as FBC and imply a dual function for the protein. The amplification of the HRAS or the SMARCB1 loci in BRCA2 males has not been reported in previous MBC studies. The relevance of this finding is uncertain, but as SMARCB1 is a tumour suppressor gene, it may be a bystander effect and may again reiterate the strength of BRCA2 drive in MBCs. This is also supported, perhaps, by the strong correlation seen between copy number changes in tumour suppressor genes PTEN, ATM, RB1, SMAD4 and STK11 (r>0.64), but less so with TP53 (only with PTEN, ATM and RB1) within the BRCAX cohort but not within BRCA2, suggesting alternate drivers between these groups.
A genome-wide association study of the germline of 823 MBC patients identified 17 SNPs mapping to six independent genomic regions that were associated with predisposition to MBC (Orr et al, 2012). However, none of these are present on our panel. A substantial proportion of our cases were included in the above study and our findings are in keeping with previous findings in that no candidate variants within our gene panel are suggestive of MBC predisposition or are of substantial clinical relevance. More so, variant frequency appears similar across all BRCA subgroups, suggesting the absence of at least a strong modifier of BRCA affect.
Conclusion
This the first study to perform high-throughput somatic sequencing on familial MBC. It shows differences between BRCA2 and BRCAX tumours, with the former harbouring TP53 mutations and the latter containing frequent PIK3CA mutations similar to luminal A FBCs. Overall, mutation frequency was lower than that seen in FBC.
Analysis of gene copy number analysis also showed differences between BRCA2 and BRCAX cohorts. While some gains and losses were similar to that reported previously in both MBC and FBC, we have identified specific gains that are particular to BRCA2 tumours. Comparison of coexpressed genes also demonstrated differences between BRCA2 and BRCAX cases with a distinct concordance of tumour suppressor genes with BRCAX patients and more heterogeneity in BRCA2 cases. We also noted more gene losses than other previous MBC studies, suggesting that familial MBCs may be a unique cohort among which difference exist between BRCA2 and BRCAX cancers. Furthermore, from a future treatment perspective, the findings suggest that different pathways may be screened and targeted depending on the BRCA status of MBC patients.
Change history
09 December 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Anelli A, Anelli TF, Youngson B, Rosen PP, Borgen PI (1995) Mutations of the p53 gene in male breast cancer. Cancer 75: 2233–2238.
Benvenuti S, Frattini M, Arena S, Zanon C, Cappelletti V, Coradini D, Daidone MG, Pilotti S, Pierotti MA, Bardelli A (2008) PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat 29: 284–288.
Bloom KJ, Govil H, Gattuso P, Reddy V, Francescatti D (2001) Status of HER-2 in male and female breast carcinoma. Am J Surg 182: 389–392.
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70.
Cleton-Jansen AM, Collins N, Lakhani SR, Weissenbach J, Devilee P, Cornelisse CJ, Stratton MR (1995) Loss of heterozygosity in sporadic breast tumours at the BRCA2 locus on chromosome 13q12–q13. Br J Cancer 72: 1241–1244.
Corso G, Marrelli D, Pascale V, Vindigni C, Roviello F (2012) Frequency of CDH1 germline mutations in gastric carcinoma coming from high- and low-risk areas: metanalysis and systematic review of the literature. BMC Cancer 12: 8.
Dawson PJ, Schroer KR, Wolman SR (1996) Ras and p53 genes in male breast cancer. Mod Pathol 9: 367–370.
Deb S, Do H, Byrne D, Jene N, Dobrovic A, Fox SB (2013) PIK3CA mutations are frequently observed in BRCAX but not BRCA2 -associated male breast cancer. Breast Cancer Res 15: R69.
Deb S, Jene N, Fox SB (2012a) Genotypic and phenotypic analysis of familial male breast cancer shows under representation of the HER2 and basal subtypes in BRCA-associated carcinomas. BMC Cancer 12: 510.
Deb S, Jene N, Kconfab I, Fox SB (2012b) Genotypic and phenotypic analysis of male breast cancer shows under representation of the HER2 and basal subtypes in BRCA-associated carcinomas. BMC Cancer 12: 510.
Do H, Wong SQ, Li J, Dobrovic A (2013) Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem 59: 1376–1383.
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868.
Giordano SH, Buzdar AU, Hortobagyi GN (2002) Breast cancer in men. Ann Intern Med 137: 678–687.
Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD (2001) TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res 61: 4092–4097.
Harvey JM, Clark GM, Osborne CK, Allred DC (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17 (5): 1474.
Hiort O, Naber SP, Lehners A, Muletta-Feurer S, Sinnecker GH, Zollner A, Komminoth P (1996) The role of androgen receptor gene mutations in male breast carcinoma. J Clin Endocrinol Metab 81: 3404–3407.
Johansson I, Nilsson C, Berglund P, Strand C, Jonsson G, Staaf J, Ringner M, Nevanlinna H, Barkardottir RB, Borg A, Olsson H, Luts L, Fjallskog ML, Hedenfalk I (2011) High-resolution genomic profiling of male breast cancer reveals differences hidden behind the similarities with female breast cancer. Breast Cancer Res Treat 129: 747–760.
Korde LA, Zujewski JA, Kamin L, Giordano S, Domchek S, Anderson WF, Bartlett JM, Gelmon K, Nahleh Z, Bergh J, Cutuli B, Pruneri G, Mccaskill-stevens W, Gralow J, Hortobagyi G, Cardoso F (2010) Multidisciplinary meeting on male breast cancer: summary and research recommendations. J Clin Oncol 28: 2114–2122.
Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, Marchionni L, van Diest PJ (2012) Oncogene amplification in male breast cancer: analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res Treat 135: 49–58.
Kwiatkowska E, Teresiak M, Breborowicz D, Mackiewicz A (2002) Somatic mutations in the BRCA2 gene and high frequency of allelic loss of BRCA2 in sporadic male breast cancer. Int J Cancer 98: 943–945.
Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, Tothill RW, Halgamuge SK, Campbell IG, Gorringe KL (2012) CONTRA: copy number analysis for targeted resequencing. Bioinformatics 28: 1307–1313.
Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, Malone KE, Daling JR, Delrow JJ, Trask BJ, Hsu L, Porter PL (2004) Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res 64: 8541–8549.
Loughrey M, Provan PJ, Byth K, Balleine RL (2008) Histopathological features of ‘BRCAX' familial breast cancers in the kConFab resource. Pathology 40: 352–358.
Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MC, Dunn SE, Hayes M, van de Rijn M, Bajdik C, Gilks CB (2004) Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res 10: 6143–6151.
Muir D, Kanthan R, Kanthan SC (2003) Male versus female breast cancers. A population-based comparative immunohistochemical analysis. Arch Pathol Lab Med 127: 36–41.
Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374.
Orr N, Lemnrau A, Cooke R, Fletcher O, Tomczyk K, Jones M, Johnson N, Lord CJ, Mitsopoulos C, Zvelebil M, Mcdade SS, Buck G, Blancher C, Trainer AH, James PA, Bojesen SE, Bokmand S, Nevanlinna H, Mattson J, Friedman E, Laitman Y, Palli D, Masala G, Zanna I, Ottini L, Giannini G, Hollestelle A, Ouweland AM, Novakovic S, Krajc M, Gago-Dominguez M, Castelao JE, Olsson H, Hedenfalk I, Easton DF, Pharoah PD, Dunning AM, Bishop DT, Neuhausen SL, Steele L, Houlston RS, Garcia-Closas M, Ashworth A, Swerdlow AJ (2012) Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet 44: 1182–1184.
Ottini L, Masala G, D’amico C, Mancini B, Saieva C, Aceto G, Gestri D, Vezzosi V, Falchetti M, de Marco M, Paglierani M, Cama A, Bianchi S, Mariani-Costantini R, Palli D (2003) BRCA1 and BRCA2 mutation status and tumor characteristics in male breast cancer: a population-based study in Italy. Cancer Res 63: 342–347.
Rudlowski C, Schulten HJ, Golas MM, Sander B, Barwing R, Palandt JE, Schlehe B, Lindenfelser R, Moll R, Liersch T, Schumpelick V, Gunawan B, Fuzesi L (2006) Comparative genomic hybridization analysis on male breast cancer. Int J Cancer 118: 2455–2460.
Tirkkonen M, Kainu T, Loman N, Johannsson OT, Olsson H, Barkardottir RB, Kallioniemi OP, Borg A (1999) Somatic genetic alterations in BRCA2-associated and sporadic male breast cancer. Genes Chromosomes Cancer 24: 56–61.
Tommasi S, Mangia A, Iannelli G, Chiarappa P, Rossi E, Ottini L, Mottolese M, Zoli W, Zuffardi O, Paradiso A (2011) Gene copy number variation in male breast cancer by aCGH. Cell Oncol (Dordr) 34: 467–473.
van de Rijn M, Gilks CB (2004) Applications of microarrays to histopathology. Histopathology 44: 97–108.
Weiss JR, Moysich KB, Swede H (2005) Epidemiology of male breast cancer. Cancer Epidemiol Biomarkers Prev 14: 20–26.
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF American Society of Clinical Oncology; College of American Pathologists (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25 (1): 118–145.
Acknowledgements
We thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow-up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia No. 628333) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. Siddhartha Deb received a postgraduate scholarship from the NHMRCS. Stephen Fox and Alexander Dobrovic received funding from the National Breast Cancer Foundation.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Deb, S., Wong, S., Li, J. et al. Mutational profiling of familial male breast cancers reveals similarities with luminal A female breast cancer with rare TP53 mutations. Br J Cancer 111, 2351–2360 (2014). https://doi.org/10.1038/bjc.2014.511
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.511
Keywords
This article is cited by
-
The Association of PTEN Gene Mutations with the Breast Cancer Risk: A Systematic Review and Meta-analysis
Biochemical Genetics (2023)
-
Male and female breast cancer: the two faces of the same genetic susceptibility coin
Breast Cancer Research and Treatment (2021)
-
Management of Rare Breast Tumors and Male Breast Cancer
Current Breast Cancer Reports (2017)
-
Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer
Tumor Biology (2016)