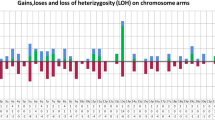
Abstract
Genetic events mediating transformation from premalignant monoclonal gammopathies (MG) to multiple myeloma (MM) are unknown. To obtain a comprehensive genomic profile of MG from the early to late stages, we performed high-resolution analysis of purified plasma cells from 20 MGUS, 20 smoldering MM (SMM) and 34 MM by high-density 6.0 SNP array. A progressive increase in the incidence of copy number abnormalities (CNA) from MGUS to SMM and to MM (median 5, 7.5 and 12 per case, respectively) was observed (P=0.006). Gains on 1q, 3p, 6p, 9p, 11q, 19p, 19q and 21q along with 1p, 16q and 22q deletions were significantly less frequent in MGUS than in MM. Although 11q and 21q gains together with 16q and 22q deletions were apparently exclusive of MM status, we observed that these abnormalities were also present in minor subclones in MGUS. Overall, a total of 65 copy number-neutral LOH (CNN-LOH) were detected. Their frequency was higher in active MM than in the asymptomatic entities (P=0.047). A strong association between genetic lesions and fragile sites was also detected. In summary, our study shows an increasing genomic complexity from MGUS to MM and identifies new chromosomal regions involved in CNA and CNN-LOH.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 2009; 113: 5412–5417.
Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM . A monoclonal gammopathy precedes multiple myeloma in most patients. Blood 2009; 113: 5418–5422.
Kyle RA, Greipp PR . Smoldering multiple myeloma. N Engl J Med 1980; 302: 1347–1349.
Perez-Persona E, Vidriales MB, Mateo G, Garcia-Sanz R, Mateos MV, de Coca AG et al. New criteria to identify risk of progression in monoclonal gammopathy of uncertain significance and smoldering multiple myeloma based on multiparameter flow cytometry analysis of bone marrow plasma cells. Blood 2007; 110: 2586–2592.
Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF et al. A Long-Term Study of Prognosis in Monoclonal Gammopathy of Undetermined Significance. N Engl J Med 2002; 346: 564–569.
Kyle RA, Remstein ED, Therneau TM, Dispenzieri A, Kurtin PJ, Hodnefield JM et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med 2007; 356: 2582–2590.
Davies FE, Dring AM, Li C, Rawstron AC, Shammas MA, O'Connor SM et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood 2003; 102: 4504–4511.
Hallek M, Bergsagel PL, Anderson KC . Multiple myeloma: increasing evidence for a multistep transformation process. Blood 1998; 91: 3–21.
Avet-Loiseau H, Li JY, Morineau N, Facon T, Brigaudeau C, Harousseau JL et al. Monosomy 13 Is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Blood 1999; 94: 2583–2589.
Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust A et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 2002; 100: 1417–1424.
Kaufmann H, Ackermann J, Baldia C, Nosslinger T, Wieser R, Seidl S et al. Both IGH translocations and chromosome 13q deletions are early events in monoclonal gammopathy of undetermined significance and do not evolve during transition to multiple myeloma. Leukemia 2004; 18: 1879–1882.
Lopez-Corral L, Gutierrez NC, Vidriales MB, Mateos MV, Rasillo A, Garcia-Sanz R et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin Cancer Res 2011; 17: 1692–1700.
Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010; 116: e56–e65.
Leone PE, Walker BA, Jenner MW, Chiecchio L, Dagrada G, Protheroe RK et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res 2008; 14: 6033–6041.
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471: 467–472.
International Myeloma Working Group Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol 121: 749–757 2010.
Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW . Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res 2006; 115: 205–214.
Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood 2007; 109: 1202–1210.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST . American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011; 13: 680–685.
Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2007; 21: 143–150.
Debacker K, Kooy RF . Fragile sites and human disease. Hum Mol Genet 2007; 16: R150–R158.
Dillon LW, Burrow AA, Wang YH . DNA instability at chromosomal fragile sites in cancer. Curr Genomics 2010; 11: 326–337.
Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood 1999; 93: 4365–4374.
Monni O, Oinonen R, Elonen E, Franssila K, Teerenhovi L, Joensuu H et al. Gain of 3q and deletion of 11q22 are frequent aberrations in mantle cell lymphoma. Genes Chromosomes Cancer 1998; 21: 298–307.
Knuutila S, Autio K, Aalto Y . Online access to CGH data of DNA sequence copy number changes. Am J Pathol 2000; 157: 689.
Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G . Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer 1999; 25: 195–204.
Magrangeas F, vet-Loiseau H, Munshi NC, Minvielle S . Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 2011; 118: 675–678.
Cheng SH, Ng MH, Lau KM, Liu HS, Chan JC, Hui AB et al. 4q loss is potentially an important genetic event in MM tumorigenesis: identification of a tumor suppressor gene regulated by promoter methylation at 4q13.3, platelet factor 4. Blood 2007; 109: 2089–2099.
Rasillo A, Tabernero MD, Sanchez ML, Perez de AM, Martin AM, Hernandez J et al. Fluorescence in situ hybridization analysis of aneuploidization patterns in monoclonal gammopathy of undetermined significance versus multiple myeloma and plasma cell leukemia. Cancer 2003; 97: 601–609.
Bozec A, Bakiri L, Hoebertz A, Eferl R, Schilling AF, Komnenovic V et al. Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature 2008; 454: 221–225.
Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem 2011; 286: 38095–38102.
Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci USA 2009; 106: 5831–5836.
Weitzman JB, Fiette L, Matsuo K, Yaniv M . JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell 2000; 6: 1109–1119.
Schepers H, Wierenga AT, van GD, Eggen BJ, Vellenga E, Schuringa JJ . Reintroduction of C/EBPalpha in leukemic CD34+ stem/progenitor cells impairs self-renewal and partially restores myelopoiesis. Blood 2007; 110: 1317–1325.
Thoennissen NH, Thoennissen GB, Abbassi S, Nabavi-Nouis S, Sauer T, Doan NB et al. Transcription factor CCAAT/enhancer-binding protein alpha and critical circadian clock downstream target gene PER2 are highly deregulated in diffuse large B-cell lymphoma. Leuk Lymphoma 2012; 21 Feb.
Fuchs O, Provaznikova D, Kocova M, Kostecka A, Cvekova P, Neuwirtova R et al. CEBPA polymorphisms and mutations in patients with acute myeloid leukemia, myelodysplastic syndrome, multiple myeloma and non-Hodgkin's lymphoma. Blood Cells Mol Dis 2008; 40: 401–405.
Rossi EA, Rossi DL, Cardillo TM, Stein R, Goldenberg DM, Chang CH . Preclinical studies on targeted delivery of multiple IFNalpha2b to HLA-DR in diverse hematologic cancers. Blood 2011; 118: 1877–1884.
Cheng I, Levin AM, Tai YC, Plummer S, Chen GK, Neslund-Dudas C et al. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer 2012; 51: 66–76.
Johnstone RW, Frew AJ, Smyth MJ . The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008; 8: 782–798.
Jourdan M, Reme T, Goldschmidt H, Fiol G, Pantesco V, De VJ et al. Gene expression of anti- and pro-apoptotic proteins in malignant and normal plasma cells. Br J Haematol 2009; 145: 45–58.
Ripperger T, von NN, Kamphues K, Emura M, Lehmann U, Tauscher M et al. Promoter methylation of PARG1, a novel candidate tumor suppressor gene in mantle-cell lymphomas. Haematologica 2007; 92: 460–468.
Wu X, Shi J, Wu Y, Tao Y, Hou J, Meng X et al. Arsenic trioxide-mediated growth inhibition of myeloma cells is associated with an extrinsic or intrinsic signaling pathway through activation of TRAIL or TRAIL receptor 2. Cancer Biol Ther 2010; 10: 1201–1214.
Leich E, Salaverria I, Bea S, Zettl A, Wright G, Moreno V et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood 2009; 114: 826–834.
Hartmann EM, Campo E, Wright G, Lenz G, Salaverria I, Jares P et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood 2010; 116: 953–961.
Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 2006; 108: 1733–1743.
Walker BA, Morgan GJ . Use of single nucleotide polymorphism-based mapping arrays to detect copy number changes and loss of heterozygosity in multiple myeloma. Clin Lymphoma Myeloma 2006; 7: 186–191.
Dickens NJ, Walker BA, Leone PE, Johnson DC, Brito JL, Zeisig A et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res 2010; 16: 1856–1864.
Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007; 12: 131–144.
Remy L, Trespeuch C . [Matrilysin-1 and cancer pathology]. Med Sci 2005; 21: 498–502.
Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG . Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J Neuropathol Exp Neurol 2011; 70: 568–577.
Barrow J, mowicz-Brice M, Cartmill M, MacArthur D, Lowe J, Robson K et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol 2011; 13: 212–222.
Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R . The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 2005; 122: 835–847.
Laczmanska I, Sasiadek MM . Tyrosine phosphatases as a superfamily of tumor suppressors in colorectal cancer. Acta Biochim Pol 2011; 58: 467–470.
Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010; 466: 941–946.
Glover TW . Common fragile sites. Cancer Lett 2006; 232: 4–12.
O'Keefe LV, Richards RI . Common chromosomal fragile sites and cancer: focus on FRA16D. Cancer Lett 2006; 232: 37–47.
Acknowledgements
This work was partially supported by Spanish FIS (PI080568 and PS0901897), the Spanish Myeloma Network Program (RD06/0020/0006) and the ‘Gerencia Regional de Salud, Junta de Castilla y León’ (GRS 702/A/11) grant. MES is supported by the Ministerio de Sanidad y Consumo (CA08/00212). We thank ‘Grupo Español de Mieloma’ clinicians for providing MM samples and S González, I Rodriguez, I Isidro, T Prieto y V Gutierrez for technical assistance.
AUTHOR CONTRIBUTIONS
LLC and MES participated in the design of the study, carried out all molecular studies, prepared the database for the final analysis and prepared the initial version of the paper. SB participated in the analysis of the study and provided preapproval of the final version of the paper. RG-S, MG and MVM participated in the design of the study and supervised the statistical analysis of the study. LAC and JMS participated in the statistical analysis of the study. EMG provided technical assistance. JB, AO, MH-G, PG and JH are clinicians who contributed to collection samples. JMH-R participated in the design of the study and provided the FISH results. JFSM and NCG were the initial persons promoting the study. They were responsible for the overall group and for the most important revision of the draft, as well as the persons who gave their final approval of the version to be published.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Rights and permissions
About this article
Cite this article
López-Corral, L., Sarasquete, M., Beà, S. et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia 26, 2521–2529 (2012). https://doi.org/10.1038/leu.2012.128
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.128
Keywords
This article is cited by
-
Structural variants shape the genomic landscape and clinical outcome of multiple myeloma
Blood Cancer Journal (2022)
-
Whole-genome sequencing reveals progressive versus stable myeloma precursor conditions as two distinct entities
Nature Communications (2021)
-
Immunogenetic characterization of clonal plasma cells in systemic light-chain amyloidosis
Leukemia (2021)
-
Monoclonal Gammopathy of Undetermined Significance: Current Concepts and Future Prospects
Current Hematologic Malignancy Reports (2020)
-
Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability
Leukemia (2019)