Abstract
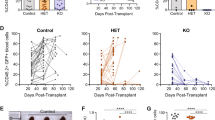
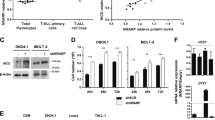
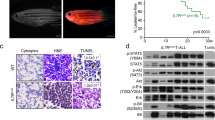
Activating NOTCH1 mutations occur in ~60% of human T-cell acute lymphoblastic leukemias (T-ALLs), and mutations disrupting the transcription factor IKZF1 (IKAROS) occur in ~5% of cases. To investigate the regulatory interplay between these driver genes, we have used a novel transgenic RNA interference mouse model to produce primary T-ALLs driven by reversible Ikaros knockdown. Restoring endogenous Ikaros expression in established T-ALL in vivo acutely represses Notch1 and its oncogenic target genes including Myc, and in multiple primary leukemias causes disease regression. In contrast, leukemias expressing high levels of endogenous or engineered forms of activated intracellular Notch1 (ICN1) resembling those found in human T-ALL rapidly relapse following Ikaros restoration, indicating that ICN1 functionally antagonizes Ikaros in established disease. Furthermore, we find that IKAROS mRNA expression is significantly reduced in a cohort of primary human T-ALL patient samples with activating NOTCH1/FBXW7 mutations, but is upregulated upon acute inhibition of aberrant NOTCH signaling across a panel of human T-ALL cell lines. These results demonstrate for the first time that aberrant NOTCH activity compromises IKAROS function in mouse and human T-ALL, and provide a potential explanation for the relative infrequency of IKAROS gene mutations in human T-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Pui C, Robison LL, Look AT . Acute lymphoblastic leukaemia. Lancet 2008; 371: 1030–1043.
Yashiro-Ohtani Y, Ohtani T, Pear WS . Notch regulation of early thymocyte development. Semin Immunol 2010; 22: 261–269.
Aster JC, Blacklow SC, Pear WS . Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol 2011; 223: 262–273.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 2007; 204: 1813–1824.
Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 2007; 204: 1825–1835.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 2006; 20: 2096–2109.
Wendorff AA, Koch U, Wunderlich FT, Wirth S, Dubey C, Brüning JC et al, Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 2010; 33: 671–684.
King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013; 153: 1552–1566.
Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood 2014; 123: 1040–1050.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 1996; 183: 2283–2291.
Beverly LJ, Capobianco AJ . Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 2003; 3: 551–564.
López-Nieva P, Santos J, Fernández-Piqueras J . Defective expression of Notch1 and Notch2 in connection to alterations of c-Myc and Ikaros in gamma-radiation-induced mouse thymic lymphomas. Carcinogenesis 2004; 25: 1299–1304.
Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 2008; 133: 727–741.
Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K . Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med 1999; 190: 1201–1214.
Yoshida T, Yao-Ming NgS, Zuniga-Pflucker JC, Georgopoulos K . Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 2006; 7: 382–391.
Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 1996; 5: 537–549.
Winandy S, Wu P, Georgopoulos K . A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 1995; 83: 289–299.
Papathanasiou P, Perkins AC, Cobb BS, Ferrini R, Sridharan R, Hoyne GF et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 2003; 19: 131–144.
Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR et al. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol 2006; 26: 209–220.
Mantha S, Ward M, McCafferty J, Herron A, Palomero T, Ferrando A et al. Activating Notch1 mutations are an early event in T-cell malignancy of Ikaros point mutant Plastic/+ mice. Leuk Res 2007; 31: 321–327.
Chari S, Winandy S . Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol 2008; 181: 6265–6274.
Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay A-S et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood 2010; 116: 5443–5454.
Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood 2010; 116: 5455–5464.
Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S . Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol Cell Biol 2005; 25: 1645–1654.
Kathrein KL, Chari S, Winandy S . Ikaros directly represses the Notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem 2008; 283: 10476–10484.
Kleinmann E, Geimer Le Lay A-S, Sellars M, Kastner P, Chan S . Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol 2008; 28: 7465–7475.
Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F et al. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity 2010; 33: 685–698.
Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 2012; 13: 86–94.
Geimer Le Lay AS, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal 2014; 7: ra28.
Marcais A, Jeannet R, Hernandez L, Soulier J, Sigaux F, Chan S et al. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res 2010; 34: 426–429.
Kastner P, Chan S . Role of Ikaros in T-cell acute lymphoblastic leukemia. World J Biol Chem 2011; 2: 108–114.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 2011; 145: 145–158.
Liao Y, Smyth GK, Shi W . The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013; 41: e108.
Robinson MD, McCarthy DJ, Smyth GK . edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43 (in press).
Law CW, Chen Y, Shi W, Smyth GK . Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29.
Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 2014; 15: 283–293.
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008; 322: 1377–1380.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–480.
Schmitt TM, Zuniga-Pflucker JC . Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 2002; 17: 749–756.
Liu GJ, Cimmino L, Jude JG, Hu Y, Witkowski MT, McKenzie MD et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev 2014; 28: 1337–1350.
Kim WI, Wiesner SM, Largaespada DA . Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol 2007; 35: 1231–1239.
Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood 2013; 122: 74–82.
Ma S, Pathak S, Mandal M, Trinh L, Clark M, Lu R . Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol 2010; 30: 4149–4158.
Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 2014; 20: 1130–1137.
Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci USA 2014; 111: E4946–E4953.
Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A et al. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol 2004; 172: 5230–5239.
Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210.
Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 2011; 208: 1809–1822.
Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 2012; 18: 298–301.
Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell 2012; 48: 445–458.
Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK . ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 2010; 26: 2176–2182.
Acknowledgements
We thank Mathew Salzone, Melanie Salzone, M Dayton, E Lanera, G Dabrowski, P Kennedy, K Stoev, C Smith, L Wilkins, S Brown and WEHI Bioservices staff for mouse work; W Alexander and E Major for ES cell and mouse resources; R Lane, J Corbin and A Keniry for technical assistance; E Viney and J Sarkis at the Australian Phenomics Network Transgenic RNAi service; M Everest and M Tinning at the Australian Genome Research Facility; and W Shi for assistance with exactSNP. We also thank the Children’s Oncology Group for primary human T-ALL samples, S Lowe and J Zuber for vectors and D Largaespada for Vav-tTA mice, and also S Lowe, J Zuber, D Izon, N Kershaw and members of the Dickins laboratory for advice and discussions. This work was supported by the National Health and Medical Research Council of Australia Project Grants 575535 and 1024599, Program Grant 490037, Senior Research Fellowship (GKS), Career Development Fellowship (RAD) and Early Career Fellowship (LC). IA was supported by the National Institutes of Health (1RO1CA133379, 1RO1CA105129, 1RO1CA149655, 5RO1CA173636 and 5RO1CA169784), the William Lawrence and Blanche Hughes Foundation, The Leukemia & Lymphoma Society, The V Foundation for Cancer Research and the St Baldrick’s Foundation. The work was also funded by Australian Government NHMRC IRIISS, an Australian Research Council Future Fellowship (SLN), Boehringer Ingelheim (MB), an ERC Advanced Grant (291740-LymphoControl) from the European Community’s Seventh Framework Program (MB), the Leukaemia Foundation of Australia (scholarship to MW, fellowship to MDM), a Sylvia and Charles Viertel Charitable Foundation Fellowship (RAD), Victorian State Government OIS grants and a Victorian Endowment for Science, Knowledge and Innovation (VESKI) Fellowship (RAD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Witkowski, M., Cimmino, L., Hu, Y. et al. Activated Notch counteracts Ikaros tumor suppression in mouse and human T-cell acute lymphoblastic leukemia. Leukemia 29, 1301–1311 (2015). https://doi.org/10.1038/leu.2015.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.27