Abstract
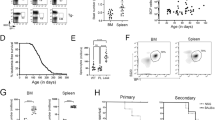
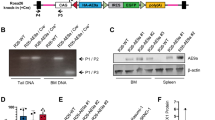
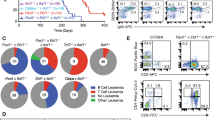
The CALM/AF10 fusion gene is found in various hematological malignancies including acute myeloid leukemia (AML), T-cell acute lymphoblastic leukemia and malignant lymphoma. We have previously identified the leukemia stem cell (LSC) in a CALM/AF10-driven murine bone marrow transplant AML model as B220+ lymphoid cells with B-cell characteristics. To identify the target cell for leukemic transformation or ‘cell of origin of leukemia’ (COL) in non-disturbed steady-state hematopoiesis, we inserted the CALM/AF10 fusion gene preceded by a loxP-flanked transcriptional stop cassette into the Rosa26 locus. Vav-Cre-induced panhematopoietic expression of the CALM/AF10 fusion gene led to acute leukemia with a median latency of 12 months. Mice expressing CALM/AF10 in the B-lymphoid compartment using Mb1-Cre or CD19-Cre inducer lines did not develop leukemia. Leukemias had a predominantly myeloid phenotype but showed coexpression of the B-cell marker B220, and had clonal B-cell receptor rearrangements. Using whole-exome sequencing, we identified an average of two to three additional mutations per leukemia, including activating mutations in known oncogenes such as FLT3 and PTPN11. Our results show that the COL for CALM/AF10 leukemia is a stem or early progenitor cell and not a cell of B-cell lineage with a phenotype similar to that of the LSC in CALM/AF10+ leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK . The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA 1996; 93: 4804–4809.
Dreyling MH, Schrader K, Fonatsch C, Schlegelberger B, Haase D, Schoch C et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood 1998; 91: 4662–4667.
Carlson KM, Vignon C, Bohlander S, Martinez-Climent JA, Le Beau MM, Rowley JD . Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia 2000; 14: 100–104.
Jones LK, Chaplin T, Shankar A, Neat M, Patel N, Samuel DP et al. Identification and molecular characterisation of a CALM-AF10 fusion in acute megakaryoblastic leukaemia. Leukemia 2001; 15: 910–914.
Abdou SM, Jadayel DM, Min T, Swansbury GJ, Dainton MG, Jafer O et al. Incidence of MLL rearrangement in acute myeloid leukemia, and a CALM-AF10 fusion in M4 type acute myeloblastic leukemia. Leuk Lymphoma 2002; 43: 89–95.
Narita M, Shimizu K, Hayashi Y, Taki T, Taniwaki M, Hosoda F et al. Consistent detection of CALM-AF10 chimaeric transcripts in haematological malignancies with t(10;11)(p13;q14) and identification of novel transcripts. Br J Haematol 1999; 105: 928–937.
Nakamura F, Maki K, Arai Y, Nakamura Y, Mitani K . Monocytic leukemia with CALM/AF10 rearrangement showing mediastinal emphysema. Am J Hematol 2003; 72: 138–142.
Salmon-Nguyen F, Busson M, Daniel M, Leblanc T, Bernard OA, Berger R . CALM-AF10 fusion gene in leukemias: simple and inversion-associated translocation (10;11). Cancer Genet Cytogenet 2000; 122: 137–140.
Kumon K, Kobayashi H, Maseki N, Sakashita A, Sakurai M, Tanizawa A et al. Mixed-lineage leukemia with t(10;11)(p13;q21): an analysis of AF10-CALM and CALM-AF10 fusion mRNAs and clinical features. Genes Chromosomes Cancer 1999; 25: 33–39.
Silliman CC, McGavran L, Wei Q, Miller LA, Li S, Hunger SP . Alternative splicing in wild-type AF10 and CALM cDNAs and in AF10-CALM and CALM-AF10 fusion cDNAs produced by the t(10;11)(p13–14;q14–q21) suggests a potential role for truncated AF10 polypeptides. Leukemia 1998; 12: 1404–1410.
Kobayashi H, Hosoda F, Maseki N, Sakurai M, Imashuku S, Ohki M et al. Hematologic malignancies with the t(10;11) (p13;q21) have the same molecular event and a variety of morphologic or immunologic phenotypes. Genes Chromosomes Cancer 1997; 20: 253–259.
Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood 2003; 102: 1000–1006.
Tebar F, Bohlander SK, Sorkin A . Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell 1999; 10: 2687–2702.
Klebig ML, Wall MD, Potter MD, Rowe EL, Carpenter DA, Rinchik EM . Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc Natl Acad Sci USA 2003; 100: 8360–8365.
Chaplin T, Ayton P, Bernard OA, Saha V, Della Valle V, Hillion J et al. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood 1995; 85: 1435–1441.
Aasland R, Gibson TJ, Stewart AF . The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 1995; 20: 56–59.
Saha V, Chaplin T, Gregorini A, Ayton P, Young BD . The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc Natl Acad Sci USA 1995; 92: 9737–9741.
Deshpande AJ, Rouhi A, Lin Y, Stadler C, Greif PA, Arseni N et al. The clathrin-binding domain of CALM and the OM-LZ domain of AF10 are sufficient to induce acute myeloid leukemia in mice. Leukemia 2011; 25: 1718–1727.
Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y . Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol 2006; 8: 1017–1024.
Greif PA, Tizazu B, Krause A, Kremmer E, Bohlander SK . The leukemogenic CALM/AF10 fusion protein alters the subcellular localization of the lymphoid regulator Ikaros. Oncogene 2008; 27: 2886–2896.
Caudell D, Zhang Z, Chung YJ, Aplan PD . Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res 2007; 67: 8022–8031.
Caudell D, Harper DP, Novak RL, Pierce RM, Slape C, Wolff L et al. Retroviral insertional mutagenesis identifies Zeb2 activation as a novel leukemogenic collaborating event in CALM-AF10 transgenic mice. Blood 2010; 115: 1194–1203.
Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell 2006; 10: 363–374.
Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 2002; 34: 251–256.
Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA 2006; 103: 13789–13794.
Rickert RC, Roes J, Rajewsky K . B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 1997; 25: 1317–1318.
Morse HC III, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 2002; 100: 246–258.
Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 2002; 100: 238–245.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
R Development Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2008.
Gregory R, Warnes BB, Bonebakker L, Gentleman R, Huber A, Liaw W et algplots: Various R programming tools for plotting data. R package version 2.14.1, 2014. Available at: http://CRAN.R-project.org/package=gplot(accessed December 2014).
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Buick RN, Till JE, McCulloch EA . Colony assay for proliferative blast cells circulating in myeloblastic leukaemia. Lancet 1977; 1: 862–863.
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994; 367: 645–648.
Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006; 442: 818–822.
Bonnet D, Dick JE . Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–737.
Kovacic B, Hoelbl A, Litos G, Alacakaptan M, Schuster C, Fischhuber KM et al. Diverging fates of cells of origin in acute and chronic leukaemia. EMBO Mol Med 2012; 4: 283–297.
Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell 2011; 20: 39–52.
Duque-Afonso J, Feng J, Scherer F, Lin CH, Wong SH, Wang Z et al. Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J Clin Invest 2015; 125: 3667–3680.
Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia 2005; 19: 1948–1957.
Novak RL, Harper DP, Caudell D, Slape C, Beachy SH, Aplan PD . Gene expression profiling and candidate gene resequencing identifies pathways and mutations important for malignant transformation caused by leukemogenic fusion genes. Exp Hematol 2012; 40: 1016–1027.
Small D . FLT3 mutations: biology and treatment. Hematology Am Soc Hematol Educ Program 2006;, 178–184.
Kratz CP, Niemeyer CM, Castleberry RP, Cetin M, Bergsträsser E, Emanuel PD et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood 2005; 106: 2183–2185.
Sutterlin C, Polishchuk R, Pecot M, Malhotra V . The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell 2005; 16: 3211–3222.
Zhu X, Morales FC, Agarwal NK, Dogruluk T, Gagea M, Georgescu MM . Moesin is a glioma progression marker that induces proliferation and Wnt/beta-catenin pathway activation via interaction with CD44. Cancer Res 2013; 73: 1142–1155.
Clucas J, Valderrama F . ERM proteins in cancer progression. J Cell Sci 2014; 127 (Part 2): 267–275.
Parry M, Rose-Zerilli MJ, Gibson J, Ennis S, Walewska R, Forster J et al. Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PLoS One 2013; 8: e83244.
Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood 2009; 114: 651–658.
Mulaw MA, Krause A, Deshpande AJ, Krause LF, Rouhi A, La Starza R et al. CALM/AF10-positive leukemias show upregulation of genes involved in chromatin assembly and DNA repair processes and of genes adjacent to the breakpoint at 10p12. Leukemia 2012; 26: 1012–1019.
Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 2010; 7: 174–185.
Greaves MF, Chan LC, Furley AJ, Watt SM, Molgaard HV . Lineage promiscuity in hemopoietic differentiation and leukemia. Blood 1986; 67: 1–11.
So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML . MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell 2003; 3: 161–171.
Acknowledgements
We thank Frank Constantini from Columbia University, USA, for kindly providing the pBigT and Rosa26PA vectors, to Dimitris Kioussis from MRC National Institute for Medical Research, UK, and Matthias Kieslinger from Helmholtz Zentrum München, Germany, for the Vav-Cre mouse line, to Elias Hobeika and Michael Reth, both from MPI Freiburg, Germany, for the mb1Cre mouse line (referred to as Mb1-Cre in this manuscript), and to Klaus Rajewsky from MDC Berlin, Germany, for the CD19-Cre mouse line. This work was supported by Deutsche Forschungsgemeinschaft Grants to SKB, EW, KS and UZ (SFB 684 A6, A12 and A13). SKB and MC are supported by Leukemia and Blood Cancer New Zealand and the family of Marijanna Kumerich.
Author contributions
SD and SKB designed the experiments. SD and BT performed experiments. AK, MRS and MD generated the knock-in R26LSLCA mouse line, and EW supervised this work. SV and PAG analyzed WES data. TH analyzed GEP data. BK performed the FACS sorting. LQM performed histology and IHC. SKB, AG and HB operated the sequencing platform. SK performed the sequencing experiments. KS contributed to the manuscript. UZS supervised the animal experiments. AV, KM and MR-T performed and analyzed the gene panel sequencing. SD, MC and SKB analyzed data and wrote the manuscript. SKB supervised the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Dutta, S., Krause, A., Vosberg, S. et al. The target cell of transformation is distinct from the leukemia stem cell in murine CALM/AF10 leukemia models. Leukemia 30, 1166–1176 (2016). https://doi.org/10.1038/leu.2015.349
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.349