Abstract
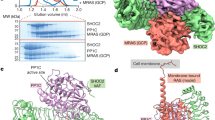
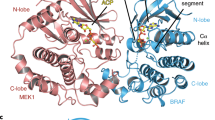
In metazoans, the Ras–Raf–MEK (mitogen-activated protein-kinase kinase)–ERK (extracellular signal-regulated kinase) signalling pathway relays extracellular stimuli to elicit changes in cellular function and gene expression. Aberrant activation of this pathway through oncogenic mutations is responsible for a large proportion of human cancer. Kinase suppressor of Ras (KSR)1,2,3 functions as an essential scaffolding protein to coordinate the assembly of Raf–MEK–ERK complexes4,5. Here we integrate structural and biochemical studies to understand how KSR promotes stimulatory Raf phosphorylation of MEK (refs 6, 7). We show, from the crystal structure of the kinase domain of human KSR2 (KSR2(KD)) in complex with rabbit MEK1, that interactions between KSR2(KD) and MEK1 are mediated by their respective activation segments and C-lobe αG helices. Analogous to BRAF (refs 8, 9), KSR2 self-associates through a side-to-side interface involving Arg 718, a residue identified in a genetic screen as a suppressor of Ras signalling1,2,3. ATP is bound to the KSR2(KD) catalytic site, and we demonstrate KSR2 kinase activity towards MEK1 by in vitro assays and chemical genetics. In the KSR2(KD)–MEK1 complex, the activation segments of both kinases are mutually constrained, and KSR2 adopts an inactive conformation. BRAF allosterically stimulates the kinase activity of KSR2, which is dependent on formation of a side-to-side KSR2–BRAF heterodimer. Furthermore, KSR2–BRAF heterodimerization results in an increase of BRAF-induced MEK phosphorylation via the KSR2-mediated relay of a signal from BRAF to release the activation segment of MEK for phosphorylation. We propose that KSR interacts with a regulatory Raf molecule in cis to induce a conformational switch of MEK, facilitating MEK’s phosphorylation by a separate catalytic Raf molecule in trans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kornfeld, K., Hom, D. B. & Horvitz, H. R. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans . Cell 83, 903–913 (1995)
Sundaram, M. & Han, M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83, 889–901 (1995)
Therrien, M. et al. KSR, a novel protein kinase required for RAS signal transduction. Cell 83, 879–888 (1995)
Clapéron, A. & Therrien, M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene 26, 3143–3158 (2007)
Kolch, W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nature Rev. Mol. Cell Biol. 6, 827–837 (2005)
Therrien, M., Michaud, N. R., Rubin, G. M. & Morrison, D. K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 10, 2684–2695 (1996)
Michaud, N. R. et al. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl Acad. Sci. USA 94, 12792–12796 (1997)
Rajakulendran, T., Sahmi, M., Lefrancois, M., Sicheri, F. & Therrien, M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009)
Wan, P. T. et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004)
Ohren, J. F. et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nature Struct. Mol. Biol. 11, 1192–1197 (2004)
Cai, Z., Chehab, N. H. & Pavletich, N. P. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol. Cell 35, 818–829 (2009)
Fischmann, T. O. et al. Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry 48, 2661–2674 (2009)
Iverson, C. et al. RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer. Cancer Res. 69, 6839–6847 (2009)
Denouel-Galy, A. et al. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr. Biol. 8, 46–55 (1998)
Yu, W., Fantl, W. J., Harrowe, G. & Williams, L. T. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr. Biol. 8, 56–64 (1998)
Stewart, S. et al. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 19, 5523–5534 (1999)
McKay, M. M., Ritt, D. A. & Morrison, D. K. Signaling dynamics of the KSR1 scaffold complex. Proc. Natl Acad. Sci. USA 106, 11022–11027 (2009)
Muller, J., Cacace, A. M., Lyons, W. E., McGill, C. B. & Morrison, D. K. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell. Biol. 20, 5529–5539 (2000)
Zhang, Y. et al. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89, 63–72 (1997)
Sugimoto, T., Stewart, S., Han, M. & Guan, K. L. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 17, 1717–1727 (1998)
Xu, W., Harrison, S. C. & Eck, M. J. Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 (1997)
Douziech, M., Sahmi, M., Laberge, G. & Therrien, M. A. KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila . Genes Dev. 20, 807–819 (2006)
Jura, N., Shan, Y., Cao, X., Shaw, D. E. & Kuriyan, J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl Acad. Sci. USA 106, 21608–21613 (2009)
Shi, F., Telesco, S. E., Liu, Y., Radhakrishnan, R. & Lemmon, M. A. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl Acad. Sci. USA 107, 7692–7697 (2010)
Alessi, D. R., Cuenda, A., Cohen, P., Dudley, D. T. & Saltiel, A. R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo . J. Biol. Chem. 270, 27489–27494 (1995)
Statsuk, A. V. et al. Tuning a three-component reaction for trapping kinase substrate complexes. J. Am. Chem. Soc. 130, 17568–17574 (2008)
Roy, F., Laberge, G., Douziech, M., Ferland-McCollough, D. & Therrien, M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16, 427–438 (2002)
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007)
Collaborative Computer Project 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Claude, J. B., Suhre, K., Notredame, C., Claverie, J. M. & Abergel, C. CaspR: a web server for automated molecular replacement using homology modelling. Nucleic Acids Res. 32, W606–W609 (2004)
Poirot, O., Suhre, K., Abergel, C., O’Toole, E. & Notredame, C. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment Nucleic Acids Res. 32, W37–W40 (2004)
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
McGuffin, L. J. & Jones, D. T. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics 19, 874–881 (2003)
Cronin, C. N., Lim, K. B. & Rogers, J. Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells. Protein Sci. 16, 2023–2029 (2007)
Allen, J. J. et al. A semisynthetic epitope for kinase substrates. Nature Methods 4, 511–516 (2007)
Bankston, D. et al. A scaleable synthesis of BAY 43-9006: A potent raf kinase inhibitor for the treatment of cancer. Org. Process Res. Dev. 6, 777–781 (2002)
Hertz, N. T. et al. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr. Prot. . Chem. Biol. 2, 15–36 (2010)
Trinidad, J. C. et al. Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 (2008)
Acknowledgements
This work was supported by a Cancer Research UK grant to D.B., ICR studentships to D.F.B. and W.C.H.C. and HHMI grant to K.M.S. We thank staff at the ESRF for help with data collection and K. Wood and V. Good for help with protein production and Z. Zhang for assistance with cloning. Mass spectrometry was made possible by NIH grants NCRR RR015804 and NCRR RR001614. The MEK1/p50Cdc37 baculovirus was a gift from C. Vaughan. We thank Cell Signaling Technologies for help with phosphospecific MEK antibodies.
Author information
Authors and Affiliations
Contributions
D.F.B. determined and analysed the MEK1–KSR2 structure; A.C.D. conducted biochemical analysis of Raf–KSR–MEK phosphorylation and inhibitor studies; N.T.H. carried out phosphoproteomics mass spectrometry studies; W.C.H.C. helped with protein production; A.L.B. analysed mass spectrometry data; K.M.S. designed and analysed experiments relating to the Raf–KSR–MEK phosphorylation and inhibitor studies; and D.B. designed experiments and analysed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
This file contains Supplementary Figures 1-17 with legends, Supplementary Tables 1-2 and additional references. (PDF 26560 kb)
Rights and permissions
About this article
Cite this article
Brennan, D., Dar, A., Hertz, N. et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 (2011). https://doi.org/10.1038/nature09860
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09860
This article is cited by
-
KSR2-14–3-3ζ complex serves as a biomarker and potential therapeutic target in sorafenib-resistant hepatocellular carcinoma
Biomarker Research (2022)
-
Qualitative differences in disease-associated MEK mutants reveal molecular signatures and aberrant signaling-crosstalk in cancer
Nature Communications (2022)
-
Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues
Nature Communications (2020)
-
Genome-wide screening identifies novel genes implicated in cellular sensitivity to BRAFV600E expression
Oncogene (2020)
-
Structural basis for the action of the drug trametinib at KSR-bound MEK
Nature (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.