Abstract
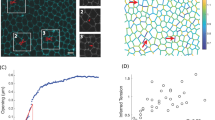
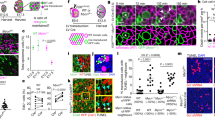
The development and maintenance of an epithelium requires finely balanced rates of growth and cell death. However, the mechanical and biochemical mechanisms that ensure proper feedback control of tissue growth1,2,3,4, which when deregulated contribute to tumorigenesis, are poorly understood. Here we use the fly notum as a model system5 to identify a novel process of crowding-induced cell delamination that balances growth to ensure the development of well-ordered cell packing. In crowded regions of the tissue, a proportion of cells undergo a serial loss of cell–cell junctions and a progressive loss of apical area, before being squeezed out by their neighbours. This path of delamination is recapitulated by a simple computational model of epithelial mechanics, in which stochastic cell loss relieves overcrowding as the system tends towards equilibrium. We show that this process of delamination is mechanistically distinct from apoptosis-mediated cell extrusion6,7,8 and precedes the first signs of cell death. Overall, this analysis reveals a simple mechanism that buffers epithelia against variations in growth. Because live-cell delamination constitutes a mechanistic link between epithelial hyperplasia and cell invasion, this is likely to have important implications for our understanding of the early stages of cancer development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hufnagel, L., Teleman, A. A., Rouault, H., Cohen, S. M. & Shraiman, B. I. On the mechanism of wing size determination in fly development. Proc. Natl Acad. Sci. USA 104, 3835–3840 (2007)
Kafer, J., Hayashi, T., Maree, A. F., Carthew, R. W. & Graner, F. Cell adhesion and cortex contractility determine cell patterning in the Drosophila retina. Proc. Natl Acad. Sci. USA 104, 18549–18554 (2007)
Farhadifar, R., Roper, J. C., Aigouy, B., Eaton, S. & Julicher, F. The influence of cell mechanics, cell–cell interactions, and proliferation on epithelial packing. Curr. Biol. 17, 2095–2104 (2007)
Aegerter-Wilmsen, T. et al. Exploring the effects of mechanical feedback on epithelial topology. Development 137, 499–506 (2010)
Cohen, M., Georgiou, M., Stevenson, N. L., Miodownik, M. & Baum, B. Dynamic filopodia transmit intermittent Delta–Notch signaling to drive pattern refinement during lateral inhibition. Dev. Cell 19, 78–89 (2010)
Rosenblatt, J., Raff, M. C. & Cramer, L. P. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11, 1847–1857 (2001)
Toyama, Y., Peralta, X. G., Wells, A. R., Kiehart, D. P. & Edwards, G. S. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science 321, 1683–1686 (2008)
Manjon, C., Sanchez-Herrero, E. & Suzanne, M. Sharp boundaries of Dpp signalling trigger local cell death required for Drosophila leg morphogenesis. Nature Cell Biol. 9, 57–63 (2007)
Zeitlinger, J. & Bohmann, D. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126, 3947–3956 (1999)
Fristrom, D. The cellular basis of epithelial morphogenesis. A review. Tissue Cell 20, 645–690 (1988)
Irvine, K. D. & Wieschaus, E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841 (1994)
Brand, A. H., Manoukian, A. S. & Perrimon, N. Ectopic expression in Drosophila . Methods Cell Biol. 44, 635–654 (1994)
Koto, A., Kuranaga, E. & Miura, M. Apoptosis ensures spacing pattern formation of Drosophila sensory organs. Curr. Biol. 21, 278–287 (2011)
Emoto, Y. Cellular aggregation facilitates anoikis in MDCK cells. J. Physiol. Sci. 58, 371–380 (2008)
Oda, H. & Tsukita, S. Real-time imaging of cell–cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J. Cell Sci. 114, 493–501 (2001)
Leevers, S. J., Weinkove, D., MacDougall, L. K., Hafen, E. & Waterfield, M. D. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15, 6584–6594 (1996)
Morin, X., Daneman, R., Zavortink, M. & Chia, W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila . Proc. Natl Acad. Sci. USA 98, 15050–15055 (2001)
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature 448, 151–156 (2007)
Georgiou, M., Marinari, E., Burden, J. & Baum, B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18, 1631–1638 (2008)
Acknowledgements
We thank J. Rosenblatt for communicating results before publication; N. Tapon, F. Pichaud, G. Charras and J. Rohn for helpful comments on the text; and the members of the Baum laboratory, in particular K. Van Hegan, J. Bellis and J. Beira. B.B. and E.M. thank Cancer Research UK for funding. B.B. also thanks University College London, Wellcome and the Royal Society for financial support.
Author information
Authors and Affiliations
Contributions
E.M. conducted the experiments detailed in the paper, aided by S.C. The laser ablation work was done with technical assistance from J.G. A.M. performed the theoretical analysis. T.D. oversaw the theoretical analysis. B.B. oversaw the experimental work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Table 1 and Supplementary Figures 1-13. (PDF 14326 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP. The movie covers the period 14-26h AP, during which cells delaminate as indicated in Figure 1. Total time is 740 minutes, with frames at 10 minute intervals. (MOV 1626 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP, expressing p110 PI3K driven by the pnr-Gal4 driver. Cells delaminate during this time interval (indicated in Figure 2). Time interval between frames is 10 minutes and the video length is 650 minutes. (MOV 7618 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP, expressing Tsc1 and Tsc2 driven by the pnr-Gal4 driver. Cells delaminate during this time interval (indicated in Figure 2). Time interval between acquired frames is 10 minutes and the video length is 670 minutes. (MOV 1029 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP, expressing dsRNA targeting Rbp5S driven from the pnr-Gal4 driver. Cells delaminate during this time interval. Time interval between frames is 10 minutes and the video length is 770 minutes. (MOV 3134 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP, expressing dsRNA targeting Cdc25 driven from the pnr-Gal4 driver. Cells do not divide but delaminate during this time interval (indicated in Figure S3). Time interval between acquired frames is 5 minutes and the video length is 135 minutes. (MOV 124 kb)
Supplementary Movie
This movie shows developing notum visualized with E-Cadherin::GFP, expressing DIAP1 driven from the pnr-Gal4 driver. Cells delaminate during this time interval (indicated in Fig. 4). Time interval between frames is 10 minutes and the video length is 730 minutes. (MOV 1767 kb)
Supplementary Movie
The movie shows the simulation of tissue evolution with parameters corresponding to the wildtype midline tissue, when the geometry is elongated and the tissue is compressed (γ=2). It corresponds to the ‘wildtype’ case in Figure 3B. Cells are shaded according to the number of neighbors – lighter shades correspond to fewer neighbours. (MOV 1423 kb)
Rights and permissions
About this article
Cite this article
Marinari, E., Mehonic, A., Curran, S. et al. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 542–545 (2012). https://doi.org/10.1038/nature10984
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10984
This article is cited by
-
Homeostatic regulation of renewing tissue cell populations via crowding control: stability, robustness and quasi-dedifferentiation
Journal of Mathematical Biology (2024)
-
Neuronal migration prevents spatial competition in retinal morphogenesis
Nature (2023)
-
Microtubule disassembly by caspases is an important rate-limiting step of cell extrusion
Nature Communications (2022)
-
Sculpting tissues by phase transitions
Nature Communications (2022)
-
Actomyosin controls planarity and folding of epithelia in response to compression
Nature Materials (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.