Abstract
2-Oxoglutarate (2OG)-dependent oxygenases have important roles in the regulation of gene expression via demethylation of N-methylated chromatin components1,2 and in the hydroxylation of transcription factors3 and splicing factor proteins4. Recently, 2OG-dependent oxygenases that catalyse hydroxylation of transfer RNA5,6,7 and ribosomal proteins8 have been shown to be important in translation relating to cellular growth, TH17-cell differentiation and translational accuracy9,10,11,12. The finding that ribosomal oxygenases (ROXs) occur in organisms ranging from prokaryotes to humans8 raises questions as to their structural and evolutionary relationships. In Escherichia coli, YcfD catalyses arginine hydroxylation in the ribosomal protein L16; in humans, MYC-induced nuclear antigen (MINA53; also known as MINA) and nucleolar protein 66 (NO66) catalyse histidine hydroxylation in the ribosomal proteins RPL27A and RPL8, respectively. The functional assignments of ROXs open therapeutic possibilities via either ROX inhibition or targeting of differentially modified ribosomes. Despite differences in the residue and protein selectivities of prokaryotic and eukaryotic ROXs, comparison of the crystal structures of E. coli YcfD and Rhodothermus marinus YcfD with those of human MINA53 and NO66 reveals highly conserved folds and novel dimerization modes defining a new structural subfamily of 2OG-dependent oxygenases. ROX structures with and without their substrates support their functional assignments as hydroxylases but not demethylases, and reveal how the subfamily has evolved to catalyse the hydroxylation of different residue side chains of ribosomal proteins. Comparison of ROX crystal structures with those of other JmjC-domain-containing hydroxylases, including the hypoxia-inducible factor asparaginyl hydroxylase FIH and histone Nε-methyl lysine demethylases, identifies branch points in 2OG-dependent oxygenase evolution and distinguishes between JmjC-containing hydroxylases and demethylases catalysing modifications of translational and transcriptional machinery. The structures reveal that new protein hydroxylation activities can evolve by changing the coordination position from which the iron-bound substrate-oxidizing species reacts. This coordination flexibility has probably contributed to the evolution of the wide range of reactions catalysed by oxygenases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klose, R. J. & Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nature Rev. Mol. Cell Biol. 8, 307–318 (2007)
Walport, L. J., Hopkinson, R. J. & Schofield, C. J. Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534 (2012)
Kaelin, W. G., Jr & Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008)
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009)
Fu, Y. et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Edn Engl. 49, 8885–8888 (2010)
Kato, M. et al. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 39, 1576–1585 (2011)
van den Born, E. et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nature Commun. 2, 172 (2011)
Ge, W. et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nature Chem. Biol. 8, 960–962 (2012)
Feng, T. et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell 53, 645–654 (2014)
Henri, J. et al. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J. Biol. Chem. 285, 30767–30778 (2010)
Loenarz, C. et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl Acad. Sci. USA 111, 4019–4024 (2014)
Yosef, N. et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013)
Teichmann, M., Dumay-Odelot, H. & Fribourg, S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 3, 2–7 (2012)
Hausinger, R. P. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 21–68 (2004)
Clifton, I. J. et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 100, 644–669 (2006)
Szilágyi, A. & Zavodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8, 493–504 (2000)
Elkins, J. M. et al. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278, 1802–1806 (2003)
Del Rizzo, P. A., Krishnan, S. & Trievel, R. C. Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol. Cell. Biol. 32, 4044–4052 (2012)
Yang, C. G. et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 (2008)
Woon, E. C. et al. Linking of 2-oxoglutarate and substrate binding sites enables potent and highly selective inhibition of JmjC histone demethylases. Angew. Chem. Int. Edn Engl. 51, 1631–1634 (2012)
Horton, J. R. et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nature Struct. Mol. Biol. 17, 38–43 (2010)
Ng, S. S. et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448, 87–91 (2007)
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012)
Sengoku, T. & Yokoyama, S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 25, 2266–2277 (2011)
Sinha, K. M., Yasuda, H., Coombes, M. M., Dent, S. Y. & de Crombrugghe, B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 29, 68–79 (2010)
Yang, M. et al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 278, 1086–1097 (2011)
Hoffart, L. M., Barr, E. W., Guyer, R. B., Bollinger, J. M., Jr & Krebs, C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc. Natl Acad. Sci. USA 103, 14738–14743 (2006)
Wong, S. D. et al. Elucidation of the Fe(IV) = O intermediate in the catalytic cycle of the halogenase SyrB2. Nature 499, 320–323 (2013)
Yang, M. et al. Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor. Angew. Chem. Int. Edn Engl. 52, 1700–1704 (2013)
Iyer, L. M., Abhiman, S., de Souza, R. F. & Aravind, L. Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 38, 5261–5279 (2010)
Guerrero, S. A., Hecht, H. J., Hofmann, B., Biebl, H. & Singh, M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl. Microbiol. Biotechnol. 56, 718–723 (2001)
Miller, R. et al. On the application of the minimal principle to solve unknown structures. Science 259, 1430–1433 (1993)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Terwilliger, T. C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 59, 38–44 (2003)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Chowdhury, R. et al. Selective small molecule probes for the hypoxia inducible factor (HIF) prolyl hydroxylases. ACS Chem. Biol. 8, 1488–1496 (2013)
Bond, C. S. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19, 311–312 (2003)
Cockman, M. E. et al. Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH). Proc. Natl Acad. Sci. USA 103, 14767–14772 (2006)
Tao, Y. et al. Structural insights into histone demethylase NO66 in interaction with osteoblast specific transcription factor Osterix and gene repression. J. Biol. Chem. 288, 16430–16437 (2013)
Lancaster, D. E. et al. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem. J. 383, 429–437 (2004)
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Regni, C. A. et al. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 28, 1953–1964 (2009)
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009)
Wang, S., Engohang-Ndong, J. & Smith, I. Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis. Biochemistry 46, 14751–14761 (2007)
Groisman, E. A. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835–1842 (2001)
Aik, W., McDonough, M. A., Thalhammer, A., Chowdhury, R. & Schofield, C. J. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struct. Biol. 22, 691–700 (2012)
McDonough, M. A., Loenarz, C., Chowdhury, R., Clifton, I. J. & Schofield, C. J. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 20, 659–672 (2010)
Han, M. V. & Zmasek, C. M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10, 356 (2009)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Acknowledgements
We thank the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, European Research Council, Medical Research Council, Oxford NIHR Biomedical Research Unit, Cancer Research UK, Arthritis Research UK, Bayer Healthcare, the Rosetree Foundation and the Slovenian Academy of Sciences and Arts (R.S.) for funding. We thank the scientists of beamlines X10SA (Swiss Light Source) and I02, I03, I04, I04-1 (Diamond Light Source) for assistance. The Structural Genomics Consortium is a registered charity (number 1097737) funded by Abbvie, Boehringer Ingelheim, the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Eli Lilly, Genome Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, Janssen, Novartis Research Foundation, Pfizer, Takeda and the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
R.C., R.S., N.C.B., C.-h.H., W.G., N.J.K., C.P., S.S.N. and E.S.P. cloned the constructs and purified proteins; R.C. and R.S. performed assays; R.C., R.S., N.C.B., S.S.N., C.-h.H. and E.S.P. crystallized the protein–ligand/substrate complexes; R.C., N.C.B., T.K., S.S.N., I.J.C., G.C.F., K.L.K., F.v.D. and M.A.M. collected and processed X-ray data; R.C., N.C.B., T.K., J.R.C.M., M.V. and M.A.M. solved and refined complex structures; R.C., R.S., M.A.M. and C.J.S. analysed data; R.C., U.O., A.J.D. and C.J.S. designed the studies; R.C. and C.J.S. wrote the paper with the help of others. See Supplementary Table 5 for further details.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Schematic protein topologies of ROXs and related 2OG-dependent oxygenases.
a–f, Protein topologies of MINA53–Mn–2OG–RPL27A(32–50) (a), NO66–Mn–NOG–RPL8(205–224) (b), RmYcfD–Mn–NOG–L16(72–91) (c), FIH–Fe–NOG–HIF-1α(786–826) (PDB accession 1H2K) (d), PHF8–Fe–NOG–H3K4me3K9me2(2–25) (PDB accession 3KV4) (e) and KDM4A–Ni–NOG–H3K9me2(7–14) (PDB accession 2OX0) (f) (substrates are not shown). DSBH core elements, labelled βI–βVIII, are in green, helices in cyan, additional β-strands in red, random coils in black and the insert between the fourth and fifth β-strands in blue. Note that not all the DSBH oxygenases maintain antiparallel hydrogen-bond pairing between βII and βVII even though the ϕ/ψ angles (βII) are within the β-region of the Ramachandran plot. Figures were generated using TopDraw47.
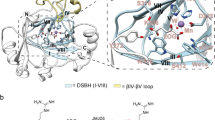
Extended Data Figure 2 ROX dimerization domains.
a, Comparison of the dimerization domains in ROXs and FIH. b, Intermolecular interactions observed at dimerization interfaces (monomer A, grey; monomer B, yellow). Validation of the functional relevance of the ROX dimers comes from biochemical and kinetic studies demonstrating loss of activities with most variants. The dimer interfaces in the ROXs are related to that of FIH; we propose that the FIH dimerization fold evolved from that of ROXs17,48. The large buried surface area (>3,000 Å2) within all ROX dimerization domains is sufficient for dimerization in solution, as reported for NO66 (ref. 49). The interactions observed in dimerization include both hydrogen bonds/electrostatic interactions and hydrophobic interactions. In the EcYcfD/RmYcfD dimerization domains, residues involved in hydrophobic interactions are mainly from α2 and are well conserved (RmYcfD residues in parentheses): Phe 214 (Met 223), Val 242 (Ile 250), Met 247 (Leu 255), Leu 250 (Ile 258), Met 253 (Leu 261), Met 254 (Leu 262), Leu 257 (Leu 265), Ile 258 (Ile 257). Hydrogen bonding/electrostatic interactions are more important in RmYcfD dimerization than in EcYcfD/human ROXs. The network of hydrogen bonds between the two RmYcfD monomers A and B includes Asp 256A–Arg 269B–Gln 259A–Asp 267B–Arg 263A, which, owing to two-fold symmetry, creates a total of eight hydrogen bonds. In EcYcfD, Leu 255 (Arg 263 in RmYcfD) is positioned at the centre of the equivalent network. Furthermore, in RmYcfD Gln 216 is positioned to hydrogen bond with the backbone amide N of Arg 234 and the carbonyl O of Leu 261. Hydrogen bonding in EcYcfD dimerization is less extensive, with only the Asn 226 amide N positioned to form a hydrogen bond to the hydroxyl group O of Thr 207 and Arg 208 hydrogen bonding with the carbonyl O of Gly 224. However, hydrophobic/aromatic clusters are involved in EcYcfD dimerization, including by the side chains of Leu 210A, Leu 223A, Tyr 217A (α1), Phe 264A, Trp 267A, Phe 268A and Phe 271A (α3) from monomer A and Val 242B, Met 247B, Leu 250B (α2) from monomer B. As in the YcfDs, in NO66 there is only one apparent salt-bridge interaction at the dimer interface, that is, between Arg 474 and Asp 495 (Arg 474A NH1–Asp 495B Oδ1, 2.9 Å; Arg 474A NH2–Asp 495B Oδ2, 2.7 Å), which links the α2 and α3 helices of opposite monomers. Similarly a ‘complex salt bridge’ is observed in MINA53 between Arg 313 and Glu 320/Asp 317 (Arg 313B NH1–Glu 320A Oε2, 3.2 Å; Arg 313B NH2–Glu 320 Oε1, 2.7 Å; Arg 313A NH2–Asp 317 Oδ1, 2.9 Å) that connects the α2 helices of different monomers. Backbone amide hydrogen bonding additionally occurs between the NO66 residues Asn 426 and Leu 454, Arg 452 and Trp 428, Phe 450 and Gly 429. MINA53 also has backbone-to-side-chain interactions between residues from flexible loops connecting α1–α2 and α2–α3 helices (Gln 297B O–Lys 331A Nζ, 3.7 Å; Ser 300B Oγ–Glu 324A O, 3.1 Å). The role of hydrophobic/aromatic clusters in dimerization is apparent in NO66 where the α2 helices from different monomers are further apart when compared with those of YcfDs and MINA53 and hence have less buried surface area. However, in NO66, an apparent hydrophobic cluster forms between the N-terminal part of α1 and the C-terminal part of α2. NO66 Trp 428 (Trp 264 in MINA53) is positioned at the start of the α1 helix of monomer A and forms the centre of a hydrophobic cluster, interacting with residues Phe 431A, Ile 435A and Leu 432A on monomer A, and Val 481B, Leu 484B, Met 462B, Phe 477B and Pro 455B on monomer B. NO66 Trp 428 also forms an apparent cation–π interaction with residue Lys 480. The similarly positioned Trp 264 in MINA53 maintains hydrophobic contacts with Phe 267 and Leu 268 of the same monomer and with Ile 290, Pro 291 and Leu 294 of the other monomer, in addition to a cation–π interaction with Arg 307. Other hydrophobic contacts observed in MINA53 dimerization involving the α1 and α2 helices of different monomers include between the side chains of residues Leu 308/α2 (interacting with Leu 319/α2 and Phe 267, Leu 268, Thr 271 of α1), Leu 312/α2 (interacting with Ile 272/α1 and Leu 315/α2) and Phe 277/α1 (interacting with Val 276, Leu 269 and Ile 272 of α1). Disruption of ROX dimerization leads to loss of activity, as observed for MINA53(R313E) and EcYcfD(I211R) variants as well as for truncated MINA53 (1–265, 1–299) without dimerization and the C-terminal domains. Non-denaturing gel electrophoresis was used to investigate ROX oligomerization states in solution, which demonstrates disruption of dimerization in EcYcfD(I211R) and MINA53(R313E). The loss of activity via destabilizing ROX dimerization is reminiscent of similar roles of FIH dimerization in catalysis (An FIH(L340R) variant that was predominantly monomeric is inactive)50. Data show mean and standard error of the mean (s.e.m.) (n = 3).
Extended Data Figure 3 Interaction of the ROX C-terminal WH domains with their respective ribosomal protein substrates.
a–e, The figure shows how ROX C-terminal domains interact with their substrates. A DALI search51 indicates that a close structural homologue of the ROX C-terminal domain is the ‘peptide clamp’ (WH) domain of MccB, an enzyme involved in the biosynthesis of the microcin C7 antibiotic52. WH domains, a subtype of the helix-turn-helix (HTH) family, are nucleic acid/protein-interacting domains and occur in different cellular pathways, from transcriptional regulation to RNA processing13. Although the overall negative charge of ROX WH domains suggests that they may not directly interact with nucleic acids, it is notable that the prokaryotic ribosomal proteins L6, which is located proximal to L16 in intact ribosomes53, and the transcriptional regulator PhoP contain WH folds54; the latter is interesting because in the E. coli K12 genome the ycfD gene is located adjacent to those for the PhoP/PhoQ two component signalling system, which is involved in stress responses55. a, General topology of the C-terminal WH domain showing two distinct binding sites for L16 (yellow) and RPL27A (magenta)/RPL8 (orange) involving residues either from an N-terminal loop connecting the WH and dimerization domains (as in RmYcfD) or from an extended loop between WH β3–β4 (as in human ROXs). b–e, Comparisons between the WH domains in MccB (b), MINA53 (c), NO66 (d) and RmYcfD/EcYcfD (e) showing the interactions observed between this domain and the substrate(s). Note that although both the RPL27A and RPL8 substrates make hydrophobic contacts with the WH domains in MINA53 (Met 405 and Met 406) (c) and NO66 (Val 576 and Tyr 577) (d), RmYcfD uses Arg 285 to form a hydrogen bond with the L16 Met 83 (RmYcfD Arg 285 NH2–L16 Met 83 O, 2.5 Å) (e). Right panels show the partial loss of activity with mutations of MINA53 (M405A), NO66 (Y577A) and EcYcfD (H277C) residues from WH domains. Data show mean and s.e.m. (n = 3).
Extended Data Figure 4 Comparison of 2OG/co-substrate binding in ROXs and representative 2OG-dependent oxygenases.
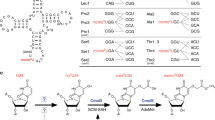
The identity of the basic residue (Arg or Lys) that binds the 2OG C5 carboyxlate via electrostatic interactions is indicated along with which of the eight DSBH (I–VIII) strands it is located on. The occurrence and positioning of the basic Arg/Lys is characteristic of each subfamily14,15. 2OG binding also involves other polar residues including alcohols, that is, a Ser (βVIII, part of the RXS motif as present in, for example, DAOCS, ANS, FTO and algal P4H) or Thr (βII, for example, as in some KDMs: JMJD3, JMJD6, PHF8 and UTX) or Tyr (non-DSBH β-strand, for example, as in FIH, KDM4A, ABH2 and PHD2) and sometimes, water molecule(s) (reviewed in refs 15,56,57). In an analogous position to the serine of the RXS motif (βVIII), human ROXs have histidine residues, MINA53 His 253/NO66 His 417 (βVIII), which form part of a hydrogen-bond network involving MINA53 Thr 255/NO66 Thr 419 (βVIII), a water molecule, and the 2OG carboxylates. Although EcYcfD/RmYcfD has Asn 197/Thr 206 at this position (βVIII), it is the conserved serine from βI (114 in EcYcfD and 122 in RmYcfD) that is positioned to hydrogen bond with the 2OG C5 carboxylate.
Extended Data Figure 5 Human ROX–substrate complexes showing disulphide crosslinking sites and difference electron density for the substrate residues.
a, Strategy adopted to obtain the crosslinked structures (the same strategy can be used for other protein hydroxylases/KDMs). b–d, Different disulphide crosslinking sites (red arrows) that form NO66–RPL8 cysteine–disulphide pairs under equilibrating conditions. Analyses of the 2OG-oxygenase–substrate complexes reveal that substrate residues at ±2 positions relative to the hydroxylated residues make interactions with enzyme residues within a ∼12 Å radius of the metal. To obtain stable NO66–RPL8 complexes, we engineered NO66 variants substituting Cys residues within ∼12 Å radius of the metal at positions considered likely to be involved in substrate binding based on the analyses of other 2OG-oxygenase–substrate structures21,22,26 and the evolutionary/phylogenetic analyses of NO66/NO66-like proteins in eukaryotes. We also substituted Cys residues at ±2 positions on the peptide substrate sequence, relative to the hydroxylated residue. Electrospray ionization–mass spectrometry (ESI–MS) assays were used to identify the best crosslinking yields for the NO66–RPL8 pairs under equilibrating conditions. The following crosslinked pairs were used for crystallization: wild-type NO66 with RPL8(G220C), a double NO66 variant L299C/C300S with RPL8(G220C), and a single NO66 variant S373C with RPL8(G214C). Structures were obtained for wild-type NO66–RPL8(G220C) (complex 1; b), NO66(L299C/C300S)–RPL8(G220C) (complex 2; c), and NO66(S373C)–RPL8(G214C) (complex 3; d) in combination with NOG/Mn(II) in C2 space group, 2.25–2.50 Å resolution with two molecules per asymmetric unit; RPL8 residues 215–223 (complex 1), 213–223 (complex 2) and 212–223 (complex 3) were observed bound to the NO66 active site. e, Superimposition of the three complex structures. Note that the key RPL8 residues (215–219), including the hydroxylated His 216, are observed in near identical conformations (r.m.s.d. 0.29–0.36 Å for Cα atoms); the similarity of the substrate positions in all the three NO66 structures suggests that they all probably represent functional complexes. On the basis of the NO66–RPL8 structures, we identified a MINA53 residue, Y209C, suitable for crosslinking, which we crystallized in complex with RPL27A(G37C) (g). Fo − Fc omit electron-density maps contoured at 3σ are shown as green (RPL8) and grey (RPL27A) meshes around the substrate residues. To test whether the wild-type/mutant enzymes and altered substrates still function catalytically we carried out endpoint and time-course assays using variable enzyme-to-substrate ratios. f, h, The biochemical data show that for both wild-type NO66 (f) and MINA53 (h) (wild type and Y209C), all the Cys-substituted peptides function as substrates. In the case of MINA53, the Y209C variant with which we obtained the MINA53–RPL27A complex structure is approximately fourfold more active than wild-type MINA53. Data are mean and s.e.m. (n = 3). We also tested wild-type NO66 for reaction between enzyme cysteines and the cysteines of modified substrate peptides by ESI–MS. Despite testing multiple combinations, we only observed disulphide formation in cases where we were also able to obtain crystal structures for substrate complexes. All possible combinations of human ROX wild type or variants and the peptides containing Cys at variable positions were used for the cross-reactivity tests: NO66: wild type, R297C, L299C/C300S, S373C, S421C; RPL8: wild type, G214C, H218C and G220C; MINA53: wild type and Y209C; RPL27A: wild type and G37C. The combined activity and MS analyses suggest that in order to form stable/crystallizable cross-linked complexes, the substrates need to be recognized by the enzyme active sites in a catalytically relevant manner (a).
Extended Data Figure 6 Mutagenesis analyses of the substrate-binding residues located on the JmjC catalytic domains of MINA53, NO66 and RmYcfD.
a–c, MINA53 (a), NO66 (b) and RmYcfD (c) are shown in colour-coded sticks. Left panels show views from the active sites of ROX–substrate complexes and the right panels show the effects of mutations on ROX catalysis. Data are mean and s.e.m. (n = 3). Analyses of ROX–substrate complexes reveal important interactions between ROX and their ribosomal protein substrates. With human ROXs, the binding of ribosomal RPL27A His 39 (light blue)/RPL8 His 216 (orange) involves a series of hydrogen bonds to backbone amides and the side chains of MINA53/NO66 residues: MINA53 Gln 136/NO66 Arg 297, MINA53 Asn 165/NO66 Asn 326, MINA53 Tyr 167/ NO66 Tyr 328 and MINA53 Ser 257/NO66 Ser 421. In addition, in the MINA53–RPL27A complex, Leu 38 and Arg 42 of RPL27A make hydrophobic contacts with MINA53 Leu 176 and a salt-bridge interaction with MINA53 Asp 333, respectively. We produced variants of all these residues to investigate their roles on substrate binding. The results of the endpoint assays as well as kinetic studies on the variants (right panels) show that substitution of these residues causes substantial losses of activity. c, In the case of RmYcfD, the hydroxylated residue L16 Arg 82 binds in a hydrophobic cleft lined by RmYcfD Tyr 137 and RmYcfD Met 120 side chains and hydrogen bonds to RmYcfD Asp 118 and RmYcfD Ser 208. To test the crystallographically observed binding mode, variants of RmYcfD residues (Asp 118, Met 120, Tyr 137 and Ser 208, highlighted) were prepared in EcYcfD (corresponding to Asp 110, Met 112, Tyr 129 and Ser 199, respectively). Mutagenesis studies on all ROXs support the crystallographically observed binding modes of the substrate residues. The combined biochemical and structural data also provide insights into the substrate selectivity of ROXs over other oxygenases.
Extended Data Figure 7 Conformational changes on substrate binding in ROX.
a–c, Conformational changes at the domain and residue levels in MINA53 (dark salmon and red with/without RPL27A, light blue) (a), NO66 (slate and cyan with/without RPL8, orange) (b) and RmYcfD (grey and split pea with/without L16, yellow) (c). Although the overall movement observed for the C-terminal WH domain on substrate binding is more significant in MINA53 as compared to other ROXs, the RmYcfD structures with and without substrate show marked local changes in the side chains of substrate-binding residues (see below). a, The inset highlights local changes to the active-site region in MINA53 in the presence (green sticks) or absence (yellow sticks) of substrate; MINA53 uses an acidic residue, Asp 333, located on an α-helix connecting the dimerization and WH domains, to form a catalytically important salt-bridge interaction with RPL27A Arg 42. Support for this statement comes from activity analyses on variants of both RPL27A and MINA53. We have previously reported that a mutation of Arg 42 in RPL27A to Ala results in <5% hydroxylation8. The D333A variant of MINA53 ablates hydroxylation (almost completely) of native RPL27A in all tested substrate:enzyme ratios (Extended Data Fig. 6). In the substrate-unbound form, MINA53 Asp 333 has two alternative conformations, indicating flexibility. The NO66 substrate RPL8 has an Ile 219 at the analogous position to Arg 42 of RPL27A that makes hydrophobic contacts with the Tyr 577 side chains from the WH domain of NO66 (b). In the case of RmYcfD, the substrate-interacting residues located on the βII–βIII loop (Tyr 137), the βIV–βV insert (Arg 169), the dimerization domain (Arg 212 and Glu 218) and on the loop connecting the dimerization and WH domains (Arg 284) are observed in different conformations in the structures with and without substrate, probably reflecting induced fit on substrate binding (c). Substitutions of these residues have variable effects on ROX catalyses (Extended Data Fig. 6).
Extended Data Figure 8 Comparison of YcfDs from E. coli and R. marinus.
a–d, Differences between YcfDs from E. coli (green) and R. marinus (grey) are shown. a, Superimposition of EcYcfD and RmYcfD–L16 complex structures showing crystallographically observed differences, particularly in the dimerization and βIV–βV loop regions. The βIV–βV insert is highlighted in crimson red and pink in EcYcfD and RmYcfD, respectively. b, Residue numbering is according to RmYcfD, with the EcYcfD numbering shown in brackets. Note that all of the directly identified substrate-binding residues are strictly conserved between EcYcfD and RmYcfD. However, some residues, particularly those located on the βIV–βV insert including Asp 118, Tyr 137 and Arg 212 in RmYcfD (Asp 110, Tyr 129 and Arg 203 in EcYcfD), are observed in different conformations, suggesting potential roles for these residues in catalysis. c, d, Predicted binding mode of L16 (yellow) to EcYcfD (green). A model complex of EcYcfD with Mn(II), NOG and L16 (residues Pro 77–Lys 84) was generated using EcYcfD-SeMet as the template and by comparison with RmYcfD–L16 and MINA53–RPL27A(32–50) structures. d, Surface representations of the EcYcfD–Mn–NOG–L16(77–84) complex, predicting key hydrogen-bond/polar interactions (dotted lines) with L16. The hydroxylated L16 Arg 81 is predicted to bind in a pocket defined by the Tyr 129 and Met 112 sidechains, which probably form π–cation and hydrophobic interactions with the L16 Arg 81 side chain, as observed in the RmYcfD–L16 crystal structure. The Arg 81 guanidino group is predicted to make electrostatic interactions with the EcYcfD Asp 110 carboxylate and hydrogen bonds to EcYcfD Ser 199. EcYcfD residues Asp 110, Met 112, Tyr 129 and Ser 199 were substituted to test the predicted mode of binding; the assay results are given in Extended Data Fig. 6c.
Extended Data Figure 9 Comparison of active-site chemistry of ROXs and related enzymes.
The figure compares active-site chemistry in representative 2OG-dependent oxygenases and directionality of the peptide substrate binding through the active site. Red/blue arrows indicate hydroxylation/demethylation sites. The active-site metals (Fe/Fe surrogates, Mn or Ni) are in colour-coded spheres.
Extended Data Figure 10 Phylogenetic relationships of human JmjC 2OG-dependent oxygenases.
The figure shows a parsimony tree constructed using Archaeopteryx v.0.9812 (ref. 58) from ClustalW59 aligned protein sequences of human JmjC-containing 2OG-dependent oxygenases showing that distinct branches of JmjC-containing oxygenases exist for hydroxylases (red), demethylases/hydroxylases (light green) and demethylases (blue).
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-5. (PDF 384 kb)
Rights and permissions
About this article
Cite this article
Chowdhury, R., Sekirnik, R., Brissett, N. et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 510, 422–426 (2014). https://doi.org/10.1038/nature13263
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13263
This article is cited by
-
MDIG-mediated H3K9me3 demethylation upregulates Myc by activating OTX2 and facilitates liver regeneration
Signal Transduction and Targeted Therapy (2023)
-
Recent Advances and Therapeutic Implications of 2-Oxoglutarate-Dependent Dioxygenases in Ischemic Stroke
Molecular Neurobiology (2023)
-
Four novel candidate causal variants for deficient homozygous haplotypes in Holstein cattle
Scientific Reports (2022)
-
Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5
Scientific Reports (2022)
-
Conservation of the unusual dimeric JmjC fold of JMJD7 from Drosophila melanogaster to humans
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.