Abstract
Mouse embryonic stem cells derived from the epiblast1 contribute to the somatic lineages and the germline but are excluded from the extra-embryonic tissues that are derived from the trophectoderm and the primitive endoderm2 upon reintroduction to the blastocyst. Here we report that cultures of expanded potential stem cells can be established from individual eight-cell blastomeres, and by direct conversion of mouse embryonic stem cells and induced pluripotent stem cells. Remarkably, a single expanded potential stem cell can contribute both to the embryo proper and to the trophectoderm lineages in a chimaera assay. Bona fide trophoblast stem cell lines and extra-embryonic endoderm stem cells can be directly derived from expanded potential stem cells in vitro. Molecular analyses of the epigenome and single-cell transcriptome reveal enrichment for blastomere-specific signature and a dynamic DNA methylome in expanded potential stem cells. The generation of mouse expanded potential stem cells highlights the feasibility of establishing expanded potential stem cells for other mammalian species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981)
Tam, P. P. & Rossant, J. Mouse embryonic chimeras: tools for studying mammalian development. Development 130, 6155–6163 (2003)
Saba-El-Leil, M. K. et al. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 4, 964–968 (2003)
Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009)
Li, X. et al. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell 8, 46–58 (2011)
Hemberger, M. et al. Parp1-deficiency induces differentiation of ES cells into trophoblast derivatives. Dev. Biol. 257, 371–381 (2003)
Koh, D. W. et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA 101, 17699–17704 (2004)
Loh, K. M., van Amerongen, R. & Nusse, R. Generating cellular diversity and spatial form: Wnt signaling and the evolution of multicellular animals. Dev. Cell 38, 643–655 (2016)
Kapoor, A. et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 (2014)
Shao, D. D. et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014)
Azzolin, L. et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 (2014)
Wilson, M. B., Schreiner, S. J., Choi, H. J., Kamens, J. & Smithgall, T. E. Selective pyrrolo-pyrimidine inhibitors reveal a necessary role for Src family kinases in Bcr–Abl signal transduction and oncogenesis. Oncogene 21, 8075–8088 (2002)
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009)
Wang, W. et al. Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Reports 13, 524–532 (2015)
Martello, G. et al. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491–504 (2012)
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008)
Kim, H. et al. Modulation of β-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 4, 2403 (2013)
Zimmerlin, L. et al. Tankyrase inhibition promotes a stable human naïve pluripotent state with improved functionality. Development 143, 4368–4380 (2016)
Morgani, S. M. et al. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Reports 3, 1945–1957 (2013)
Latos, P. A. & Hemberger, M. From the stem of the placental tree: trophoblast stem cells and their progeny. Development 143, 3650–3660 (2016)
Kolodziejczyk, A. A. et al. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471–485 (2015)
Marks, H. et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604 (2012)
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014)
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016)
Macfarlan, T. S. et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 (2012)
Abad, M. et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345 (2013)
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013)
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016)
Kubaczka, C. et al. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Reports 2, 232–242 (2014)
Niakan, K. K., Schrode, N., Cho, L. T. & Hadjantonakis, A. K. Derivation of extraembryonic endoderm stem (XEN) cells from mouse embryos and embryonic stem cells. Nat. Protoc. 8, 1028–1041 (2013)
Ryan, D. J., Yang, J., Lan, G. & Liu, P. Derivation and maintenance of mouse expanded potential stem cells. Protoc. Exch. http://doi.org/10.1038/protex.2017.102 (2017)
Nichols, J., Silva, J., Roode, M. & Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222 (2009)
Yang, J. et al. Quenching autofluorescence in tissue immunofluorescence. Wellcome Open Res. 2, 79 (2017)
Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998)
Schröter, C., Rué, P., Mackenzie, J. P. & Martinez Arias, A. FGF/MAPK signaling sets the switching threshold of a bistable circuit controlling cell fate decisions in embryonic stem cells. Development 142, 4205–4216 (2015)
Freyer, L. et al. A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev. Biol. 15, 38 (2015)
Ng, R. K. et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 10, 1280–1290 (2008)
Wang, L., Jin, Q., Lee, J. E., Su, I. H. & Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl Acad. Sci. USA 107, 7317–7322 (2010)
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Buecker, C. et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853 (2014)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012)
Zhang, Y. et al. Model-based analysis of ChIP–Seq (MACS). Genome Biol. 9, R137 (2008)
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016)
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010)
Shen, L., Shao, N., Liu, X. & Nestler, E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Ficz, G. et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359 (2013)
Acknowledgements
We thank colleagues of the Research Support Facility (B. Doe, S. Newman, E. Grau and others), Y. Hooks, Sequencing (N. Smerdon) and FACS core facilities (B. L. Ng and J. Graham) at the Sanger Institute, the animal facility at the Cancer Research UK Cambridge Institute, and P. Humphreys of the University of Cambridge, for technical support; S. Gerety for the fluorescence stereo microscope, J. K. Kim for informatics advice, S. Rice for help on DNA bisulfite sequencing analysis; J. Nichols, A. Martinez Arias, K. McDole and Y. Zheng for reagents; J. Thomson, E. Robertson and A. Ang for comments. We acknowledge the following funding and support: Wellcome Trust Clinical PhD Fellowship for Academic Clinicians (D.J.R.); PhD fellowship (Portuguese Foundation for Science and Technology, FCT (SFRH/BD/84964/2012)) (L.A.); Japan Society for the Promotion of Science fellowship (Y.T.); National Institutes of Health (RP-PG-0310-10002) (A.C.W.); European Molecular Biology Organization (ALTF938-2014) and Marie Sklodowska-Curie Individual Fellowship (M.A.E.-M.); Biotechnology and Biological Sciences Research Council (BB/K010867/1) and Wellcome Trust (095645/Z/11/Z) (W.R.); Bloodwise (12029), Cancer Research UK (C1163/A12765 and C1163/A21762) and Wellcome Trust core funding (SCI 097922/Z/11/Z) (B.G.); Leading Advanced Projects for Medical Innovation, Japan Agency for Medical Research and Development (H.N. and H.M.); National Health and Medical Research Council Senior Principal Research Fellowship (1110751) (P.P.L.T.); National Natural Science Foundation of China (81671579, 31370904, 30972691) and The National Key Research and Development Program (2017YFA0104500) (L.L.). P.L. thanks M. Stratton, A. Bradley, N. Copeland, N. Jenkins and J. Lupski for their encouragement in these experiments. P.L. is an affiliate faculty member of the Wellcome Trust-MRC Stem Cell Institute, University of Cambridge. The P.L. laboratory is supported by the Wellcome Trust (grant numbers 098051 and 206194).
Author information
Authors and Affiliations
Contributions
D.J.R. and W.W. developed EPSCM and derived mouse EPSC lines. J.Y. performed most of the experiments, made the final figures and edited the manuscript. J.C.-H.T performed and analysed the RNA-seq and chromatin immunoprecipitation followed by sequencing (ChIP–seq) experiments, produced the genomics figures and wrote the manuscript. G.L. performed most of the injections. H.M. performed independent EPSC injection experiments at Nakauchi laboratory. X.G., L.A., Y.Y., Z. Zhu, J.W., A.A.K. and C.W. performed genotyping, additional line derivation, FACS, sequencing and chimaera experiments. L.S.C. interpreted the histology data. F.Y. and B.F. karyotyped EPSC and ES cell lines. Z.Zho. performed confocal imaging and interpretation. M.A.E.-M. and W.R. provided DNA methylation data. M.W., Y.T. and A.C.W. performed additional experiments. X.C. analysed TSC RNA-seq data. J.B., J.W., R.R.-S., W.R., B.G., S.A.T., H.N. and X.Z. provided microinjection resources, sequencing and other support. L.L. and P.P.L.T. contributed intellectually and assisted with the revision of the manuscript. P.L. devised the concept, supervised the overall research project and prepared the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The Genome Research Limited has filed a provisional patent application that covers the derivation and maintenance of expanded potential stem cells (European patent application number 15797300.9-1402). P.L., D.J.R., J.Y., W.W. and X.G. are listed as inventors.
Additional information
Reviewer Information Nature thanks A.-K. Hadjantonakis and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Derivation and functional characterization of EPSC lines from pre-implantation embryos and by converting ES cells and iPS cells in EPSCM.
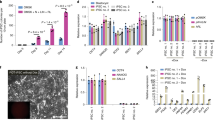
a, Phase images showing development of eight-cell mouse embryos in EPSCM or in M15. Note that in embryos in EPSCM, the blastocoel cavity was filled with cells and the embryos hatched at day 7 or 8. b, EPSCs derived from pre-implantation embryos express pluripotency markers. Analysis of pluripotency markers in two embryo-derived EPSC lines (DR25 and DR10), one reprogrammed iPS cell–EPSC line and two embryo-derived 2i/LIF ES cell lines (DR4 and DR9). Expression of pluripotency genes was relative to Gapdh in the RT–qPCR assay. Data are mean ± s.d. (n = 3). Overlaid dot plots indicate the distribution of the data. c, Expression of lineage-specific genes in the cells described in b. Expression of these genes was relative to Gapdh. Data are mean ± s.d. (n = 3). Overlaid dot plots indicate the distribution of the data. d, Spectral karyotyping of DR10-EPSCs (normal karyotype in eight out of ten metaphases) at passage 10. e, Mature teratomas from DR10-EPSCs. Left: ectodermal neural tube-like structures; middle: mesodermal cartilage; right: an endodermal gland structure of gastrointestinal type and mesodermal muscle fibres. The sections were stained with haematoxylin–eosin. f, A chimaera derived from DR10-EPSCs. g, The mCherry+ EPSCs show extensive contribution to the 14.5 d.p.c. gonad, similar to 2i/LIF ES cells. h, Additional images of EPSC contributions in the trophectoderm of the blastocyst. The two images on the top are merged live images (phase and mCherry), which show the contribution of mCherry (cytoplasmic)-labelled 2i/LIF ES cells or ES cell–EPSCs. The two at the bottom are immunostaining images to detect H2B–mCherry-expressing AB2.2 ES cell–EPSCs and the trophectoderm cells (Cdx2+). i, Rex1–GFP reporter ES cell colonies in M15, 2i/LIF or EPSCM on SNL feeder. j, Expression of the reporter GFP in ES cells cultured in EPSCM (passage 3). Top: Oct4–GFP ES cells (E14Tg2a) cultured in M15, 2i/LIF or EPSCM. In all conditions, Oct4–GFP expression was detected by FACS. Bottom: Rex1–GFP ES cells (AB2.2 background) have comparable levels of Rex1 expression (GFP) when cultured in EPSCM or subsequently returned to 2i/LIF. Negative control: wild-type ES cells. k, Expression of pluripotency genes or lineage-specific genes in ES cells cultured in EPSCM (ES cell–EPSCs). Expression of these genes was relative to Gapdh. Data are mean ± s.d. (n = 2). Overlaid dot plots indicate the distribution of the data.
Extended Data Figure 2 Characterization of EPSCs and the signalling responses.
a, Left: Oct4 distal and proximal enhancer luciferase reporter constructs. DE, the Oct4 distal enhancer; PE, the Oct4 proximal enhancer; MiniP, the minimum promoter. The Oct4 distal enhancer is active in ES cells cultured in EPSCM. Right: luciferase activities of the Oct4 DE and PE constructs were normalized to that of the MiniP construct in the same cell type. Data are mean ± s.d. (n = 3). b, Differentiation of ES cell–EPSCs in vitro into cells of three germ layers detected by the immunostaining of class III β-tubulin, α-smooth muscle actin (SMA) and Gata4. c, A male chimaera from ES cell–EPSCs, and germline transmission of the Rex1–GFP allele from the chimaera. d, Activation of X chromosomes in female ES cell–EPSCs. EPSCs or the cells differentiated from EPSCs were co-immunostained for H3K27me3 and Oct4. No discrete H3K27me3 foci were found in EPSCs. Once the cells differentiated, the foci appeared in almost all cells. Arrows point to H3K27me3 foci. e, f, Effects of small-molecule inhibitors on their respective targets. p-ERK, phosphorylated ERK; p-Src, phosphorylated Src; p-P38, phosphorylated P38; p-JNK, phosphorylated JNK. α-Tubulin was used as the loading control. Note the substantial increase of axin1. EPSCM contains 20% KSR so KSR-2i/LIF is the appropriate negative control. XAV939 in EPSCM stabilizes axin1 by inhibiting its ubiquitination. EPSCs had considerably elevated levels of axin1 compared with controls (arrow), which caused accumulation of phosphorylated β-catenin in both the cytoplasm and nuclei and decreased active β-catenin in the nucleus. α-Tubulin and histone H3 were used as the loading controls of cytoplasmic and nuclear proteins, respectively. The gel source data are in Supplementary Fig. 1. g, Reduced β-catenin–LEF/TCF activity in EPSCs detected by TOPflash luciferase assay. TOPflash luciferase activity was normalized to that of EPSCs grown in M15 medium. Data are mean ± s.d. (n = 8 for M15, 9 for N2B27-2i/LIF and EPSCM). Two-sided Student’s t-test was performed. ***P < 0.001. h, Western blot analysis shows increased p-STAT3 in ES cell–EPSCs with increased concentrations of LIF. The gel source data are in Supplementary Fig. 1. i, Upregulation of downstream LIF pathway genes in ES cell–EPSCs in response to LIF stimulation. Two-sided Student’s t-test was performed. Data are mean ± s.d. (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. j, Signalling pathway dependence in ES cell–EPSCs. ES cells and EPSCs (n = 100 for each cell type) cultured in M15, 2i/LIF or EPSCM with inhibitors of JAK (JAK inhibitor 1), FGFR (SU5402), TGFβR (A83-01) or ALK5 (SB505124). Alkaline phosphatase-positive (AP+) colonies were scored after 10 days. The JAK inhibitor substantially reduced the number of AP+ colonies from these cells. By contrast, ES cells cultured in EPSCM did not appear to be sensitive to FGFR or TGFβR inhibitors, similar to 2i/LIF ES cells. Data are mean ± s.d. (n = 3 for control, 4 for inhibitors). ***P < 0.001.
Extended Data Figure 3 Contribution of EPSCs in chimaeras.
a, Whole-mount immunostaining of 6.5 d.p.c. chimaeras for H2B–mCherry and Elf5. In the chimaeras shown, the descendants of EPSCs were found both in the embryo proper and in the ExE. One of the chimaeras (number five from the top) has EPSC contribution predominantly in the Elf-5-expressing ExE. The live images at the bottom show the contribution of EPSCs predominantly in the ExE and ectoplacental cone (EPC) (mCherry in Cambridge, GFP in Tokyo). b, Whole-mount fluorescence imaging of 14.5 d.p.c. chimaeras from morulas injected with ES cells or EPSCs. Negative control: wild-type embryo or chimaeras from unlabelled EPSCs. Em, embryo; Yo, yolk sac; Pl, placenta. Inset tables show the frequency of contribution by the stem cells to the chimaeras. c, Percentages of EPSC or ES cell contributions in independent chimaeras (n = 6 for DR10-EPSC; n = 9 for AB2.2-EPSC; n = 6 for AB2.2-ES cell control). Cells from the brain (Br), liver (Li) and placenta (Pl) were collected for flow cytometric analysis. d, Detection of polyploid placenta cells by flow cytometry. A distinct population of 8N cells is present in both mCherry+ and mCherry− placenta cells from an AB2.2-EPSC chimaera. Fetal brain cells were used as the control. No mCherry+ 8N cells could be detected in the placenta from 2i/LIF ES cells. e. Placenta sections of 14.5 d.p.c. chimaeras from either ES cells or EPSCs (AB2.2, mCherry+) were co-immunostained for mCherry and Tfap2c. mCherry+ cells were detected in the EPSC chimaera placenta; some of these cells were positively stained for both mCherry and Tfap2c (arrows). The areas framed in the low-magnification DAPI staining (insets) indicate the positions of the placenta section images. Abbreviations: laby, labyrinth; sp., spongiotrophoblast layer; gc, trophoblast giant cell layer.
Extended Data Figure 4 Contribution of EPSCs in the placenta and the yolk sac.
a, mCherry+ cells sorted from 14.5 d.p.c. EPSC chimaera placenta, cytospun to polylysine-coated slides, and co-immunostained for mCherry and trophoblast markers GCM1, ezrin and cytokeratin 7 (CK7). The arrows point to mCherry+ cells that are positive for these markers. b, mCherry+ EPSCs were injected into ROSA26–GFP–SB10 blastocysts. The chimaeras were collected at 14.5 d.p.c. mCherry+ and mCherry− placenta cells were sorted, from which genomic DNA was extracted for PCR. SB10 DNA (Sleeping Beauty transposase gene) should be amplified from the host cells, and mCherry should be amplified from the donor cells. The mCherry+ placenta cells had robust mCherry DNA amplification, but did not have any detectable host cell DNA, as no SB10 DNA amplification was found, which excludes cell fusion events between the donor cells and host cells in the placenta. By contrast, weak mCherry signal was found in mCherry− cells, which is probably because of the silencing of the CAG promoter in some placenta cells derived from the mCherry+ donor cells. Amplification of a region in the Oct4 distal enhancer region serves as the genomic DNA quality and PCR control. The gel source data are in Supplementary Fig. 1. c, Yolk sac sections of 14.5 d.p.c. chimaeras of either ES cells (panels 1–3) or EPSCs (panels 1′–3′). Chimaeras from injected Rosa26–GFP reporter EPSCs or ES cells were analysed. The original GFP fluorescence signal was quenched by fixation. A CF660C-conjugated antibody against GFP was used to detect donor cells. DAPI stains the nucleus. GFP+ cells were found in extra-embryonic mesoderm (endothelial and mesothelial cells) of the yolk sac in chimaeras produced both from EPSCs and from ES cells. In the yolk sac of EPSC chimaeras, GFP+ cells were also found in the extra-embryonic endoderm layer. Arrows point to extra-embryonic endoderm, extra-embryonic mesoderm.
Extended Data Figure 5 Contribution of a single EPSC in chimaeras.
a, Whole-mount fluorescence of 14.5 d.p.c. chimaeras from eight-cell embryos and a single EPSC or ES cell. Negative control: wild-type embryo. Inset table shows the frequency of contribution by the stem cells to the chimaeras. b, Flow cytometry analysis of mCherry+ placenta cells in single-EPSC-injected chimaeras. c, Expression of trophoblast genes in mCherry+ and mCherry− placenta cells sorted from the single-EPSC chimaera. Expression was relative to Actb and normalized to fetal brain expression. Data are mean ± s.d. (n = 3). Overlaid dot plots indicate the distribution of the data. Sorting was performed as in Fig. 2c. d, Detection of polyploid placenta cells. A distinct population of 8N cells was found both in mCherry+ and in mCherry− placenta cells in an EPSC chimaera.
Extended Data Figure 6 Minimal requirement of inhibitors in EPSCM.
a, Effects of individual inhibitors on trophectoderm contribution. Mouse ES cells (mCherry+ AB2.2) were cultured in various combinations of inhibitors for at least five passages before injection into eight-cell embryos and examination at the blastocyst stage. Blastocysts were scored for the donor cells’ trophectoderm contribution. A, A419259; X, XAV939; JNKi, JNK Inhibitor VIII; p38i, SB203580; CH, CHIR99021. All culture conditions contained KSR and LIF. For each culture condition, 38–58 embryos were scored. b, The effect of CHIR99021 (CH) in EPSCM on cells. One thousand EPSCs cultured in either EPSCM or EPSCM minus CH were plated for single-cell colony formation assay. AP+ colonies were scored on day 7 (n = 3). A two-sided Student’s t-test was performed. Data are mean ± s.d. (n = 3). ***P < 0.001. c, Gene expression in ES cells cultured in minimum sets of inhibitors. Expression of genes was relative to Gapdh in the RT–qPCR assay. Data are mean ± s.d. (n = 3). Overlaid dot plots indicate the distribution of the data. d, Flow cytometry analysis of mCherry+ placenta cells of 14.5 d.p.c. chimaeras from ES cells cultured in 2i+A medium. To minimize residual background cell sorting, a GFP channel was used to exclude autofluorescence. e, Expression of trophoblast-enriched genes in cells sorted from the placenta of chimaeras. The mCherry+ placenta cells were sorted as in d from chimaeras of ES cells cultured in 2i+A medium. Expression was relative to Actb. Data are mean ± s.d. (n = 3). Overlaid dot plots indicate the distribution of the data. f, Yolk sac sections of 14.5 d.p.c. chimaeras of ES cells (2i+A) or the wild-type negative control. Immunostaining and imaging were performed in the same way as in Extended Data Fig. 4c. The mCherry fluorescence signal was quenched by fixation. Donor cells were stained by a CF660C-conjugated antibody against mCherry (panels 1 and 1′). DAPI staining was used to detect the nucleus (panels 2 and 2′). Donor-cell derivatives were found in both the extra-embryonic mesoderm (endothelial and mesothelial cells) and extra-embryonic endoderm cells in the yolk sac of EPSC chimaera, but not in the yolk sac of the negative control chimaera (3 and 3′). Arrows point to extra-embryonic endoderm, extra-embryonic mesoderm.
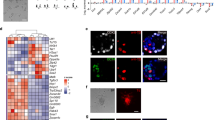
Extended Data Figure 7 Single-cell RNA-seq analyses of EPSCs.
a, Scatter plot showing the relationship of EPSC gene expression variability with expression levels in scRNA-seq data. The magenta dots represent genes that showed significantly higher variability (adjusted P < 0.1) than would be expected from the external RNA spike-ins (blue dots). b, Three-dimensional scatter plot showing the separation of 2i/LIF ES cells, M15 ES cells and EPSCs in PCA of scRNA-seq data. Each dot represents a cell. c, Bar graph showing Gene Ontology terms enriched in differentially expressed genes between 2i/LIF ES cells and EPSCs in scRNA-seq data. d, Violin plots comparing expression of selected pluripotency regulators, DNA methylation regulators and two-cell embryo-associated genes in 2i/LIF ES cells, M15 ES cells and EPSCs in scRNA-seq data. The dot marks the median of the data. EPSCs (n = 84); 2i/LIF ES cells (n = 82); M15 ES cells (n = 91). e, Box plot comparing the distribution of pre-implantation embryos, 2i/LIF ES cells and EPSCs in the first principal component in PCA. The bar and the small square in the boxes mark the median and mean scores of each group. The upper and lower hinge, and the upper and lower whiskers, mark the first and third quartiles of the data, and the smallest and largest observation excluding outliers, respectively. Outliers were determined as observations less than the first quartile −1.5× interquartile range and observations larger than the third quartile +1.5× interquartile range. The PCA was performed with differentially expressed genes between EPSCs and 2i/LIF ES cells.
Extended Data Figure 8 Comparison of EPSCs with other pluripotent stem cells and epigenomic characteristics of EPSCs.
a, Three-dimensional scatter plot showing the separation of Hex-Venus+ 2i/LIF ES cells, MERV-TdT+ ES cells, in vivo iPS cells (iviPS cell), 2i/LIF ES cells, M15 ES cells and EPSCs in PCA of bulk RNA-seq data. b, Bar graph comparing the relative expression levels of MERV transcripts in EPSCs and MERV-TdT+ ES cells to MERV-TdT− ES cells. c, Signal intensity distribution of EPSC H3K4me3 and H3K27me3 modification over gene bodies and 3 kb up- and downstream of the transcription start site (TSS) and transcription end site (TSE). Genes are classified into ‘very high’, ‘high’, ‘low’ and ‘very low’, depending on their length-corrected mean-normalized count quartiles in the single-cell RNA-seq dataset. d, Distribution of H3K27me3 (left) and H3K4me3 (right) signals at gene promoters (±3 kb from TSS). The promoters were ranked on the basis of their length-corrected mean-normalized count levels in the single-cell RNA-seq dataset. The signals were quantified as read count per million mapped reads. e, Profiles of H3K4me3 and H3K27me3 of 2i/LIF ES cells and EPSCs at selected pluripotency-associated loci. f, Bar graph showing −log(P value) of each development-related Gene Ontology term enriched in EPSC-specific bivalent genes only. g, Profiles of H3K4me3 and H3K27me3 at selected loci in pre-implantation embryos, EPSCs and 2i/LIF ES cells. Note the similar H3K27me3 profiles in EPSCs and in two-cell or eight-cell embryos. h, Profiles of H3K27ac signals of oocyte, two-cell embryos, eight-cell embryos, 2i/LIF ES cells and EPSCs in selected super-enhancer-associated genes, including the Oct4, Klf5 and Gata3 loci. Note that the super-enhancer at the Gata3 locus is present in EPSCs and in eight-cell embryos but not in 2i/LIF ES cells.
Extended Data Figure 9 Derivation and characterization of EPSC TSCs.
a, Immunofluorescence for GFP in EPSCs or 2i/LIF ES cells cultured in TX medium for 6 days. Small patches of Cdx2+ cells were detected only in differentiated EPSCs, but not 2i/LIF ES cells. DAPI stains the nucleus. The Cdx2+ cells account for around 1.0% of total cells in flow cytometry. b, Expression of Cdx2, Eomes and Tfap2c in established EPSC TSCs detected by immunostaining. Four EPSC TSC lines were established from Cdx2–GFP EPSCs. c, Heat map comparing the expression of transcription factors of pluripotency, embryo lineages or trophectoderm development in EPSCs, embryo-derived TSCs and multiple lines of EPSC-derived TSCs. The expression levels were calculated in log-transformed values of transcript per million (TPM). EPSC rep1 and rep2 are two biological replicates of EPSC RNA samples. d, DNA demethylation at the Elf5 locus in EPSC TSCs (n = 10). e, Phase images of EPSC TSCs in differentiation medium (RPMI 1640 plus 20% serum without FGF4 and heparin) for the indicated days. Arrows indicate polyploid trophoblasts. f, RT–qPCR analysis of EPSC TSCs or the control TSCs at different days of differentiation. Cdx2 was downregulated while expression of mature trophoblast genes was upregulated. Expression levels are relative to Gapdh. Data are mean ± s.d. (n = 3 technically independent samples). Overlaid dot plots indicate the distribution of the data. g, Immunofluorescence staining of differentiated EPSC TSCs or TSCs for placenta lactogen-1 (PL-1) (day 8). The insets show DAPI staining results. h, Representative images of haemorrhagic lesions in NSG mice 7 days after subcutaneous injection of EPSC TSCs or TSCs. i, Haematoxylin and eosin sections of haemorrhagic lesions of EPSC TSCs or TSCs. The images show that the well-defined haemorrhagic lesions are lined by large and occasionally multinucleated pleomorphic cells with abundant cytoplasm. These large cells that demarcate the blood-filled space are reminiscent of trophoblast giant cells.
Extended Data Figure 10 Characterization of EPSC–XEN cells and overexpression of axin1 in ES cells.
a, Live images of Gata6H2B-Venus+ XEN cells derived directly from EPSCs in vitro. b, RT–qPCR analysis of expression of cell-type-specific genes for XEN (Gata4, Gata6, Sox7 and Sox17), TSC (Cdx2) or ES cells (Nanog). Expression levels are relative to Gapdh. Data are mean ± s.d. (n = 3 technically independent samples). Two biologically independent EPSC–XEN cell lines were analysed. Overlaid dot plots indicate the distribution of the data. c, Expression of Gata4 and Gata6 in XEN cells visualized by immunostaining. d, Contribution of Gata6H2B-Venus EPSC–XEN cells in blastocysts (n = 17) and in 6.5 d.p.c. chimaeras. The donor cells in the PrE of the hatching blastocyst or in the extra-embryonic endoderm were immunostained by the antibody to Gata6. The Oct4 antibody staining marks the embryo proper. Inset table shows the contribution. e, Contribution of ES cells overexpressing axin1 in the ExE region in 6.5 d.p.c. chimaeras. Top panels, donor ES cells in the chimaera expressed axin1 under control of the Tet/On system before injection, whereas in the bottom panels the donor cells expressed axin1 from the CAG promoter. Inset table shows the contribution. f, Western blot confirming axin1 overexpression in ES cells using either the CAG promoter or the Tet/On system. The gel source data are in Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 (source gels) and the gating strategy of mCherry+ placenta cells. The mCherry+ placenta cells were analysed using LSR II Fortessa cytometry (Bector Dickinson) analyser, and the data were analysed using FlowJo. Because of the various sizes of trophoblasts in placenta, the majority of starting cells were included in FSC/SSC gates. The boundary between positive and negative is defined according the negative control. We also used a non-overlapping channel (GFP) to gate out the false positive cells due to autoflurescence in placenta cells. (PDF 1183 kb)
Supplementary Data
This file contains Bed files of signal peaks identified in EPSCs by histone modifications ChIP-seq (H3K4me3, H3K27me3 and H3K27Ac super enhancers). (XLSX 4064 kb)
Supplementary Data
This file contains a list of ChIP-seq and RNA-seq sequencing data deposited. (XLSX 53 kb)
Supplementary Data
This file contains Supplementary Tables 1-7 and a Supplementary Table guide. (ZIP 6486 kb)
Preimplantation embryo in EPSCM
Transgenic Oct4 EGFP 4C-8C embryos were cultured in a gelatinized 96 well plate in EPSCM. At 80 hours in EPSCM, the embryos were imaged on Leica AF6000 fluorescence microscope. The microscope stage and objectives were enclosed by a cage incubator and maintained at 37°C, 5% CO2. A 20X objective was used for all positions. Time-lapses were acquired using GFP filter-set and transmitted light images were sequentially captured. Images were acquired at 30 minutes interval for 55 hours in total. (MP4 19226 kb)
Source data
Rights and permissions
About this article
Cite this article
Yang, J., Ryan, D., Wang, W. et al. Establishment of mouse expanded potential stem cells. Nature 550, 393–397 (2017). https://doi.org/10.1038/nature24052
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24052
This article is cited by
-
An optimized culture system for efficient derivation of porcine expanded potential stem cells from preimplantation embryos and by reprogramming somatic cells
Nature Protocols (2024)
-
Low-input lipidomics reveals lipid metabolism remodelling during early mammalian embryo development
Nature Cell Biology (2024)
-
The rules of the totipotency treasure hunt
Nature Cell Biology (2024)
-
A new era of stem cell and developmental biology: from blastoids to synthetic embryos and beyond
Experimental & Molecular Medicine (2023)
-
Induction of mouse totipotent stem cells by a defined chemical cocktail
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.