Abstract
Expansion and remodelling of the mammary epithelium requires a tight balance between cellular proliferation, differentiation and death. To explore cell survival versus cell death decisions in this organ, we deleted the pro-survival gene Mcl-1 in the mammary epithelium. Mcl-1 was found to be essential at multiple developmental stages including morphogenesis in puberty and alveologenesis in pregnancy. Moreover, Mcl-1-deficient basal cells were virtually devoid of repopulating activity, suggesting that this gene is required for stem cell function. Profound upregulation of the Mcl-1 protein was evident in alveolar cells at the switch to lactation, and Mcl-1 deficiency impaired lactation. Interestingly, EGF was identified as one of the most highly upregulated genes on lactogenesis and inhibition of EGF or mTOR signalling markedly impaired lactation, with concomitant decreases in Mcl-1 and phosphorylated ribosomal protein S6. These data demonstrate that Mcl-1 is essential for mammopoiesis and identify EGF as a critical trigger of Mcl-1 translation to ensure survival of milk-producing alveolar cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hinck, L. & Silberstein, G. B. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 7, 245–251 (2005).
Watson, C. J. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res. 8, 203 (2006).
Kreuzaler, P. A. et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat. Cell Biol. 13, 303–309 (2011).
Czabotar, P. E., Lessene, G., Strasser, A. & Adams, J. M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15, 49–63 (2014).
Moldoveanu, T., Follis, A. V., Kriwacki, R. W. & Green, D. R. Many players in BCL-2 family affairs. Trends Biochem. Sci. 39, 101–111 (2014).
Walton, K. D. et al. Conditional deletion of the bcl-x gene from mouse mammary epithelium results in accelerated apoptosis during involution but does not compromise cell function during lactation. Mech. Dev. 109, 281–293 (2001).
Mailleux, A. A. et al. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev. Cell 12, 221–234 (2007).
Opferman, J. T. et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307, 1101–1104 (2005).
Opferman, J. T. et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426, 671–676 (2003).
Vikstrom, I. et al. Mcl-1 is essential for germinal center formation and B cell memory. Science 330, 1095–1099 (2010).
Dzhagalov, I., St John, A. & He, Y. W. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 109, 1620–1626 (2007).
Steimer, D. A. et al. Selective roles for antiapoptotic MCL-1 during granulocyte development and macrophage effector function. Blood 113, 2805–2815 (2009).
Arbour, N. et al. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J. Neurosci. 28, 6068–6078 (2008).
Malone, C. D. et al. Mcl-1 regulates the survival of adult neural precursor cells. Mol. Cell. Neurosci. 49, 439–447 (2012).
Thomas, R. L. et al. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 27, 1365–1377 (2013).
Wang, X. et al. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 27, 1351–1364 (2013).
Rios, A. C., Fu, N. Y., Lindeman, G. J. & Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014).
Ertel, F., Nguyen, M., Roulston, A. & Shore, G. C. Programming cancer cells for high expression levels of Mcl1. EMBO Rep. 14, 328–336 (2013).
Perciavalle, R. M. & Opferman, J. T. Delving deeper: MCL-1’s contributions to normal and cancer biology. Trends Cell Biol. 23, 22–29 (2013).
Thomas, L. W., Lam, C. & Edwards, S. W. Mcl-1; the molecular regulation of protein function. FEBS Lett. 584, 2981–2989 (2010).
Asselin-Labat, M. L. et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol. Cell. Biol. 31, 4609–4622 (2011).
Okamoto, T. et al. Enhanced stability of Mcl1, a prosurvival Bcl2 relative, blunts stress-induced apoptosis, causes male sterility, and promotes tumorigenesis. Proc. Natl Acad. Sci. USA 111, 261–266 (2014).
Wintermantel, T. M., Mayer, A. K., Schutz, G. & Greiner, E. F. Targeting mammary epithelial cells using a bacterial artificial chromosome. Genesis 33, 125–130 (2002).
Rudolph, M. et al. Metabolic regulation in the lactating mammary gland: a lipid synthesising machine. Physiol. Genomics 28, 323–336 (2007).
Lemay, D. G., Neville, M. C., Rudolph, M. C., Pollard, K. S. & German, J. B. Gene regulatory networks in lactation: identification of global principles using bioinformatics. BMC Syst. Biol. 1, 56 (2007).
Inuzuka, H. et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471, 104–109 (2011).
Schwickart, M. et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107 (2010).
Forster, N. et al. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev. Cell 28, 147–160 (2014).
Hynes, N. E. & Lane, H. A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354 (2005).
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Coloff, J. L. et al. Akt-dependent glucose metabolism promotes Mcl-1 synthesis to maintain cell survival and resistance to Bcl-2 inhibition. Cancer Res. 71, 5204–5213 (2011).
Mills, J. R. et al. mTORC1 promotes survival through translational control of Mcl-1. Proc. Natl Acad. Sci. USA 105, 10853–10858 (2008).
Meyuhas, O. Physiological roles of ribosomal protein S6: one of its kind. Int. Rev. Cell Mol. Biol. 268, 1–37 (2008).
Wang, D. et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81–84 (2015).
Print, C. G. et al. Apoptosis regulator bcl-w is essential for spermatogenesis but appears otherwise redundant. Proc. Natl Acad. Sci. USA 95, 12424–12431 (1998).
Luetteke, N. C. et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 126, 2739–2750 (1999).
Fowler, K. J. et al. A mutation in the epidermal growth factor receptor in waved-2 mice has a profound effect on receptor biochemistry that results in impaired lactation. Proc. Natl Acad. Sci. USA 92, 1465–1469 (1995).
Long, W. et al. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130, 5257–5268 (2003).
Muraoka-Cook, R. S., Feng, S. M., Strunk, K. E. & Earp, H. S. III ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J. Mammary Gland Biol. Neoplasia 13, 235–246 (2008).
Preuss, E., Hugle, M., Reimann, R., Schlecht, M. & Fulda, S. Pan-mammalian target of rapamycin (mTOR) inhibitor AZD8055 primes rhabdomyosarcoma cells for ABT-737-induced apoptosis by down-regulating Mcl-1 protein. J. Biol. Chem. 288, 35287–35296 (2013).
Booy, E. P., Henson, E. S. & Gibson, S. B. Epidermal growth factor regulates Mcl-1 expression through the MAPK-Elk-1 signalling pathway contributing to cell survival in breast cancer. Oncogene 30, 2367–2378 (2011).
Leu, C. M., Chang, C. & Hu, C. Epidermal growth factor (EGF) suppresses staurosporine-induced apoptosis by inducing mcl-1 via the mitogen-activated protein kinase pathway. Oncogene 19, 1665–1675 (2000).
Song, L., Coppola, D., Livingston, S., Cress, D. & Haura, E. B. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol. Ther. 4, 267–276 (2005).
Wagner, K. U. et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330 (1997).
Tarutani, M. et al. Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc. Natl Acad. Sci. USA 94, 7400–7405 (1997).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Fu, N. Y., Sukumaran, S. K., Kerk, S. Y. & Yu, V. C. Baxbeta: a constitutively active human Bax isoform that is under tight regulatory control by the proteasomal degradation mechanism. Mol. Cell 33, 15–29 (2009).
Pal, B. et al. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 3, 411–426 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Smyth, G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet Mol. Biol. 3, 1–25 (2004).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
McCarthy, D. J. & Smyth, G. K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25, 765–771 (2009).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Clancy, J. L. et al. Methods to analyze microRNA-mediated control of mRNA translation. Methods Enzymol. 431, 83–111 (2007).
Stoelzle, T., Schwarb, P., Trumpp, A. & Hynes, N. E. c-Myc affects mRNA translation, cell proliferation and progenitor cell function in the mammary gland. BMC Biol. 7, 63 (2009).
Pillai, R. S. et al. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309, 1573–1576 (2005).
Acknowledgements
We are grateful to J. Takeda and M. Takaishi for the gift of K5–cre transgenic mice and S. Glaser for provision of mice. We thank D. Huang for advice. We also thank J. L. Clancy, S. K. Archer and G. Duan for assistance with the polysome experiments, B. Helbert for genotyping, and the Animal, FACS, Imaging and Histology facilities at WEHI. This work was supported by the Australian National Health and Medical Research Council (NHMRC) grants no. 1016701, no. 1024852, no. 1086727; NHMRC IRIISS; the Victorian State Government through VCA funding of the Victorian Breast Cancer Research Consortium and Operational Infrastructure Support; and the Australian Cancer Research Foundation. N.Y.F. and A.C.R. are supported by a National Breast Cancer Foundation (NBCF)/Cure Cancer Australia Fellowship; A.T.L.L. by an Elizabeth and Vernon Puzey Scholarship from the University of Melbourne; S.A.B. by a NHMRC Postgraduate Scholarship no. 1017256; P.B., G.K.S. and G.J.L. by NHMRC Fellowships no. 1042629, no. 1058892, no. 637307; and J.E.V. by an Australia Fellowship.
Author information
Authors and Affiliations
Contributions
N.Y.F. generated and analysed all mouse models, acquired and interpreted main data sets. N.Y.F., G.J.L. and J.E.V. designed experiments. A.C.R. performed confocal microscopy imaging; B.P. qPCR analysis; A.T.L.L. and G.K.S. bioinformatic analysis; R.S. and T.P. polysome fractionation experiments; K.L. assisted with immunohistochemistry; T.B. assisted with western blot analysis; S.A.B. assisted with histological analysis; F.V. performed transplantation assays; P.B. and A.S. provided Mcl-1 mice and discussions. J.E.V. and N.Y.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Expression of Mcl-1 in the mammary gland.
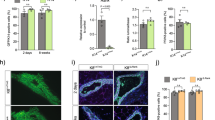
(a) Western blots showing the rapid decrease of Mcl-1 at the onset of involution. pY705 Stat3 represents a marker for involution (n = 2 independent experiments). (b) General strategy for flow cytometric analysis and sorting of mammary epithelial cells. Representative FACS plots (n = 10) showing the MaSC/basal (Lin−CD29hiCD24+) and luminal (Lin−CD29loCD24+) subsets. The luminal population was further subdivided into three distinct subpopulations based on CD14 and CD61 expression: CD14+CD61−, CD14+CD61+ and CD14−CD61−. (c) Representative FACS plots (n = 8) showing the MaSC/basal (Lin−CD29hiCD24+) and luminal (Lin−CD29loCD24+) subsets isolated from the mammary glands of 5 week-old (puberty), 10 week-old adult (virgin), 18.5 day pregnant (18.5 dP) and 2 day lactating (2 dL) mice. (d) Western blots showing the decrease in Bcl-2 levels in luminal cells that occurs during pregnancy and lactation relative to virgin glands (n = 1). (e) Representative Western blots showing Mcl-1 levels in luminal and MaSC/basal cells in puberty versus adult and 2 day lactating mammary glands, with short (SE) and long exposures (LE) indicated. (f) Quantitative RT-PCR analysis of Mcl-1 mRNA in the luminal or MaSC/basal population of mammary glands from virgin, 18.5 day pregnant (18.5 dP) or 2 day lactating (2 dL) mice. Mean ± SEM for n = 3 independent samples. (g) Western blot analysis of Bcl-2 family protein expression in the four distinct mammary epithelial cell subsets of virgin mammary glands. Approximately 250,000 cells were used for each sample (n = 1). (h) Quantitative RT-PCR analysis of Mcl-1 expression in luminal (progenitor and mature) or MaSC/basal populations isolated from virgin mammary glands. Mean ± SEM, n = 3 independent samples.
Supplementary Figure 2 Mcl-1 protein is induced in ductal and alveolar luminal cells at the switch to lactation.
Whole-mount 3D confocal images of mammary glands at 2 days of lactation (2 dL) or 18.5 days of pregnancy, showing ducts and/or alveoli. The tissues were stained for F-actin (red), Mcl-1 (green) and E-cadherin (blue) expression. An outline of the ducts is depicted in the merged images. The white arrow depicts the mesh of elongated myoepithelial cells that surround each alveolus. Representative of 3 experiments. Scale bars, 50 μm.
Supplementary Figure 3 No apparent change in the stability of Mcl-1 based on cycloheximide pulse-chase experiments and no induction of Mcl-1 by prolactin.
(a) Luminal cells (Lin−CD29loCD24+) were sorted from the mammary glands of 18.5 day pregnant or 2 day lactating mice, plated in ultra-low adherence plates and incubated with cycloheximide (CHX) for the indicated times. Lysates were subjected to western blot analysis for Mcl-1 and Actin protein levels (n = 2 experiments). (b,c) Mammary epithelial cells (Lin−CD24+) were sorted from virgin (b) or 18.5 day pregnant (c) mammary glands, starved of serum and EGF for 36 h and exposed to the lactogenic hormone prolactin for 3 days. Western blot analysis was performed to determine Mcl-1, milk and actin protein expression (n = 1).
Supplementary Figure 4 EGF is dramatically upregulated in alveolar cells at the onset of lactation.
(a) Heatmaps of the 100 most differentially expressed genes in the luminal population between late pregnancy and early lactation, clustered according to their expression values. Luminal and MaSC/basal cell populations were isolated from virgin, 18.5 day pregnant (18.5 dP) and 2 day lactating (2 dL) mammary glands (FVB/N). (c) Heatmap of gene expression for members of the EGF receptor/ligand family that are differentially expressed between late pregnancy and early lactation for the luminal population. 18.5 dP or 2 dL mice. dP, days pregnancy; dL, days lactation.
Supplementary Figure 5 Increased phospho-S6 expression in alveolar luminal cells at the onset of lactation.
(a) Immunostaining of phospho-S6 protein in the mammary glands of FVB/N mice (n = 2 mice per stage) through development. Scale bar: 50 μm. (b) 3D whole-mount confocal analysis of pS6 in the mammary glands of FVB/N mice (n = 3) at 4 days of lactation. Tissues were incubated with Phalloidin to observe the entire tissue at the cellular level. Scale bar: 50 μm. (c) Immunostaining for cleaved caspase 3 in the mammary glands isolated from mothers administrated RAD001 or vehicle at 2 days of lactation (n = 3 dams). Scale bar: 50 μm. Representative images (a–c) are shown for n = 3 experiments.
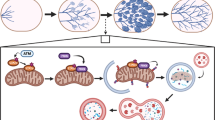
Supplementary Figure 6 Polysome profiles and RNA expression data for mammary tissue from 17 days pregnant (dP), and 2 or 4 days lactating mice (dL).
Data from two independent biological repeat experiments are shown: Experiment 1 (a–d) and Experiment 2 (e–h). (a,e) Polysome fractionation profiles (A254: absorbance at 254 nm, dominated by ribosomal RNA) from mammary gland tissue; (b,f) Representative graphs from one gradient per experiment showing RNA integrity across gradient fractions as determined on the Agilent Bioanalyzer; (c,g) Distribution of Mcl-1, Bcl-x and Actin mRNA amongst gradient fractions isolated from mammary glands at 17 dP, 2 dL and 4 dL; (d,h) Quantitative RT-PCR analysis of Mcl-1, Bcl-x and Actin mRNA steady-state levels in the unfractionated cell lysates used for polysome fractionation (mean of 3 technical replicates).
Supplementary information
Supplementary Information
Supplementary Information (PDF 1673 kb)
Supplementary Table 1
Supplementary Information (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Fu, N., Rios, A., Pal, B. et al. EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival. Nat Cell Biol 17, 365–375 (2015). https://doi.org/10.1038/ncb3117
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3117
This article is cited by
-
Effects of sustained hyperprolactinemia in late gestation on the mammary parenchymal tissue transcriptome of gilts
BMC Genomics (2023)
-
BCL-2 protein family: attractive targets for cancer therapy
Apoptosis (2023)
-
Whole genome-wide analysis of DEP family members in sheep (Ovis aries) reveals their potential roles in regulating lactation
Chemical and Biological Technologies in Agriculture (2022)
-
Impact of adaptive filtering on power and false discovery rate in RNA-seq experiments
BMC Bioinformatics (2022)
-
What can we learn from mice lacking pro-survival BCL-2 proteins to advance BH3 mimetic drugs for cancer therapy?
Cell Death & Differentiation (2022)