Abstract
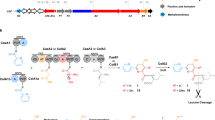
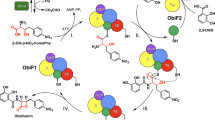
The streptothricin (ST) antibiotics, produced by Streptomyces bacteria, contain L-β-lysine ((3S)-3,6-diaminohexanoic acid) oligopeptides as pendant chains. Here we describe three unusual nonribosomal peptide synthetases (NRPSs) involved in ST biosynthesis: ORF 5 (a stand-alone adenylation (A) domain), ORF 18 (containing thiolation (T) and condensation (C) domains) and ORF 19 (a stand-alone A domain). We demonstrate that ST biosynthesis begins with adenylation of L-β-lysine by ORF 5, followed by transfer to the T domain of ORF 18. In contrast, L-β-lysine molecules adenylated by ORF 19 are used to elongate an L-β-lysine peptide chain on ORF 18, a reaction unexpectedly catalyzed by ORF 19 itself. Finally, the C domain of ORF 18 catalyzes the condensation of L-β-lysine oligopeptides covalently bound to ORF 18 with a freely diffusible intermediate to release the ST products. These results highlight an unusual activity for an A domain and unique mechanisms of crosstalk within NRPS machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Waksman, S.A. Production and activity of streptothricin. J. Bacteriol. 46, 299–310 (1943).
Ji, Z., Wang, M., Zhang, J., Wei, S. & Wu, W. Two new members of streptothricin class antibiotics from Streptomyces qinlingensis sp. nov. J. Antibiot. (Tokyo) 60, 739–744 (2007).
Kusumoto, S., Kambayashi, Y., Imaoka, S., Shima, K. & Shiba, T. Total chemical structure of streptothricin. J. Antibiot. (Tokyo) 35, 925–927 (1982).
Kusumoto, S., Imaoka, S., Kambayashi, Y. & Shiba, T. Total synthesis of antibiotic streptothricin F. Tetrahedr. Lett. 23, 2961–2964 (1982).
Hisamoto, M. et al. A-53930A and B, novel N-type Ca2+ channel blockers. J. Antibiot. (Tokyo) 51, 607–617 (1998).
Goldstein, A.L. & McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999).
Hentges, P., Van Driessche, B., Tafforeau, L., Vandenhaute, J. & Carr, A.M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22, 1013–1019 (2005).
Shen, J., Guo, W. & Kohler, J.R. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 73, 1239–1242 (2005).
Idnurm, A., Reedy, J.L., Nussbaum, J.C. & Heitman, J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3, 420–429 (2004).
Joshi, P.B., Webb, J.R., Davies, J.E. & McMaster, W.R. The gene encoding streptothricin acetyltransferase (sat) as a selectable marker for Leishmania expression vectors. Gene 156, 145–149 (1995).
Takemoto, T. et al. Physiological activity of streptothricin antibiotics. Chem. Pharm. Bull. (Tokyo) 28, 2884–2891 (1980).
Chamberlain, D.A. et al. The use of the Emu promoter with antibiotic and herbicide resistance genes for the selection of transgenic wheat callus and rice plants. Aust. J. Plant Physiol. 21, 95–112 (1994).
Hoffmann, H. et al. Pharmacokinetics of nourseothricin in laboratory animals. Arch. Exp. Veterinarmed. 40, 699–709 (1986).
Hoffmann, H. et al. Pharmacologic action profile of nourseothricin. Arch. Exp. Veterinarmed. 40, 710–720 (1986).
Härtl, A., Guttner, J., Stockel, U. & Hoffmann, H. Acute and subchronic toxicity of nourseothricin in laboratory animals. Arch. Exp. Veterinarmed. 40, 727–735 (1986).
Hamano, Y., Matsuura, N., Kitamura, M. & Takagi, H. A novel enzyme conferring streptothricin resistance alters the toxicity of streptothricin D from broad-spectrum to bacteria-specific. J. Biol. Chem. 281, 16842–16848 (2006).
Walsh, C. Antibiotics: Action, Origins, Resistance (ASM Press, 2003).
Schwarzer, D., Finking, R. & Marahiel, M.A. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20, 275–287 (2003).
Mootz, H.D., Schwarzer, D. & Marahiel, M.A. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3, 490–504 (2002).
Marahiel, M.A., Stachelhaus, T. & Mootz, H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674 (1997).
Fernández-Moreno, M.A., Vallin, C. & Malpartida, F. Streptothricin biosynthesis is catalyzed by enzymes related to nonribosomal peptide bond formation. J. Bacteriol. 179, 6929–6936 (1997).
Grammel, N. et al. A β-lysine adenylating enzyme and a β-lysine binding protein involved in poly β-lysine chain assembly in nourseothricin synthesis in Streptomyces noursei. Eur. J. Biochem. 269, 347–357 (2002).
Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W. & Huson, D.H. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808 (2005).
Zhang, W. et al. Nine enzymes are required for assembly of the pacidamycin group of peptidyl nucleoside antibiotics. J. Am. Chem. Soc. 133, 5240–5243 (2011).
Steffensky, M., Li, S.M. & Heide, L. Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroids NCIMB 11891. J. Biol. Chem. 275, 21754–21760 (2000).
Schmutz, E. et al. An unusual amide synthetase (CouL) from the coumermycin A1 biosynthetic gene cluster from Streptomyces rishiriensis DSM 40489. Eur. J. Biochem. 270, 4413–4419 (2003).
Bierman, M. et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992).
Gust, B., Challis, G.L., Fowler, K., Kieser, T. & Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100, 1541–1546 (2003).
Kieser,, T. et al. Practical Streptomyces Genetics (The John Innes Foundation, Norwich, UK, 2000).
Acknowledgements
We thank N. Iwasaki and J. Seta (Bruker Daltonics K.K.) for assistance with HPLC/Q-TOF MS data collection. Plasmids pKD46 and pCP 20 and E. coli BW25113 were supplied by H. Mori (Nara Institute of Science and Technology). This work was supported, in part, by Grant-in-Aid for Young Scientists (A) (22688007) from the Japan Society for the Promotion of Science (JSPS; to Y.H.), the Industrial Technology Research Grant Program in 2008 from the New Energy and Industrial Technology Development Organization of Japan (to Y.H.) and by Grant-in-Aid for Scientific Research on Innovative Area (23108520) from the JSPS (to Y.H.).
Author information
Authors and Affiliations
Contributions
C.M., T.U. and Y.H. conceived and designed the experiments; C.M., J.T, Y.K., M.I., M.T., K.S. and H.K. performed the experiments; and C.M. and Y.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 3658 kb)
Rights and permissions
About this article
Cite this article
Maruyama, C., Toyoda, J., Kato, Y. et al. A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat Chem Biol 8, 791–797 (2012). https://doi.org/10.1038/nchembio.1040
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1040
This article is cited by
-
History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome
npj Antimicrobials and Resistance (2024)
-
Separation of an ε-poly-l-lysine derivative by solvent extraction under a controlled interfacial potential difference
Analytical Sciences (2024)
-
Nonribosomal antibacterial peptides isolated from Streptomyces agglomeratus 5-1-3 in the Qinghai-Tibet Plateau
Microbial Cell Factories (2023)
-
N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry
Nature Communications (2023)
-
Elucidating the molecular programming of a nonlinear non-ribosomal peptide synthetase responsible for fungal siderophore biosynthesis
Nature Communications (2023)