Abstract
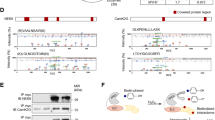
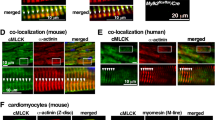
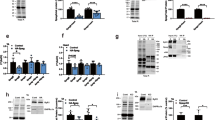
Focal adhesion kinase (FAK) regulates cellular processes that affect several aspects of development and disease. The FAK N-terminal FERM (4.1 protein–ezrin-radixin-moesin homology) domain, a compact clover-leaf structure, binds partner proteins and mediates intramolecular regulatory interactions. Combined chemical cross-linking coupled to MS, small-angle X-ray scattering, computational docking and mutational analyses showed that the FAK FERM domain has a molecular cleft (∼998 Å2) that interacts with sarcomeric myosin, resulting in FAK inhibition. Accordingly, mutations in a unique short amino acid sequence of the FERM myosin cleft, FP-1, impaired the interaction with myosin and enhanced FAK activity in cardiomyocytes. An FP-1 decoy peptide selectively inhibited myosin interaction and increased FAK activity, promoting cardiomyocyte hypertrophy through activation of the AKT–mammalian target of rapamycin pathway. Our findings uncover an inhibitory interaction between the FAK FERM domain and sarcomeric myosin that presents potential opportunities to modulate the cardiac hypertrophic response through changes in FAK activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mitra, S.K., Hanson, D.A. & Schlaepfer, D.D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 (2005).
Schaller, M.D. Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J. Cell Sci. 123, 1007–1013 (2010).
Golubovskaya, V.M., Kweh, F.A. & Cance, W.G. Focal adhesion kinase and cancer. Histol. Histopathol. 24, 503–510 (2009).
Peng, X. et al. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proc. Natl. Acad. Sci. USA 105, 6638–6643 (2008).
Clemente, C.F. et al. Targeting focal adhesion kinase with small interfering RNA prevents and reverses load-induced cardiac hypertrophy in mice. Circ. Res. 101, 1339–1348 (2007).
Luo, M. & Guan, J.L. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 289, 127–139 (2010).
DiMichele, L.A. et al. Myocyte-restricted focal adhesion kinase deletion attenuates pressure overload-induced hypertrophy. Circ. Res. 99, 636–645 (2006).
Torsoni, A.S., Constancio, S.S., Nadruz, W. Jr., Hanks, S.K. & Franchini, K.G. Focal adhesion kinase is activated and mediates the early hypertrophic response to stretch in cardiac myocytes. Circ. Res. 93, 140–147 (2003).
Dunty, J.M. & Schaller, M.D. The N termini of focal adhesion kinase family members regulate substrate phosphorylation, localization, and cell morphology. J. Biol. Chem. 277, 45644–45654 (2002).
Ceccarelli, D.F., Song, H.K., Poy, F., Schaller, M.D. & Eck, M.J. Crystal structure of the FERM domain of focal adhesion kinase. J. Biol. Chem. 281, 252–259 (2006).
Lietha, D. et al. Structural basis for the autoinhibition of focal adhesion kinase. Cell 129, 1177–1187 (2007).
Frame, M.C., Patel, H., Serrels, B., Lietha, D. & Eck, M.J. The FERM domain: organizing the structure and function of FAK. Nat. Rev. Mol. Cell Biol. 11, 802–814 (2010).
Abbi, S. et al. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol. Biol. Cell 13, 3178–3191 (2002).
Parsons, J.T. Focal adhesion kinase: the first ten years. J. Cell Sci. 116, 1409–1416 (2003).
Schaller, M.D. et al. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 14, 1680–1688 (1994).
Xing, Z. et al. Direct interaction of v-Src with the focal adhesion kinase mediated by the Src SH2 domain. Mol. Biol. Cell 5, 413–421 (1994).
Calalb, M.B., Polte, T.R. & Hanks, S.K. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell. Biol. 15, 954–963 (1995).
Kadaré, G. et al. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J. Biol. Chem. 278, 47434–47440 (2003).
Medley, Q.G. et al. Signaling between focal adhesion kinase and trio. J. Biol. Chem. 278, 13265–13270 (2003).
Chen, S.Y. & Chen, H.C. Direct interaction of focal adhesion kinase (FAK) with Met is required for FAK to promote hepatocyte growth factor-induced cell invasion. Mol. Cell. Biol. 26, 5155–5167 (2006).
Serrels, B. et al. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat. Cell Biol. 9, 1046–1056 (2007).
Pham, C.G. et al. Am. J. Physiol. Heart Circ. Physiol. 279, H2916–H2926 (2000).
Taylor, J.M., Rovin, J.D. & Parsons, J.T. A role for focal adhesion kinase in phenylephrine-induced hypertrophy of rat ventricular cardiomyocytes. J. Biol. Chem. 275, 19250–19257 (2000).
Eble, D.M. et al. Endothelin-induced cardiac myocyte hypertrophy: role for focal adhesion kinase. Am. J. Physiol. Heart Circ. Physiol. 278, H1695–H1707 (2000).
Samarel, A.M. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. Heart Circ. Physiol. 289, H2291–H2301 (2005).
Fonseca, P.M. et al. Targeting to C-terminal myosin heavy chain may explain mechanotransduction involving focal adhesion kinase in cardiac myocytes. Circ. Res. 96, 73–81 (2005).
Koch, M.H., Vachette, P. & Svergun, D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 36, 147–227 (2003).
Inagaki, K., Hahn, H.S., Dorn, G.W. II. & Mochly-Rosen, D. Additive protection of the ischemic heart ex vivo by combined treatment with delta-protein kinase C inhibitor and epsilon-protein kinase C activator. Circulation 108, 869–875 (2003).
Chen, L. et al. Opposing cardioprotective actions and parallel hypertrophic effects of delta PKC and epsilon PKC. Proc. Natl. Acad. Sci. USA 98, 11114–11119 (2001).
Marin, T.M. et al. Shp2 negatively regulates growth in cardiomyocytes by controlling focal adhesion kinase/Src and mTOR pathways. Circ. Res. 103, 813–824 (2008).
Laser, M. et al. Integrin activation and focal complex formation in cardiac hypertrophy. J. Biol. Chem. 275, 35624–35630 (2000).
Schaller, M.D., Otey, C.A., Hildebrand, J.D. & Parsons, J.T. Focal adhesion kinase and paxillin bind to peptides mimicking beta integrin cytoplasmic domains. J. Cell Biol. 130, 1181–1187 (1995).
Iglesias, A.H., Santos, L.F. & Gozzo, F.C. Identification of cross-linked peptides by high-resolution precursor ion scan. Anal. Chem. 82, 909–916 (2010).
Tovchigrechko, A. & Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 (2006).
Svergun, D.I., Barberato, C. & Koch, M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecule from atomic coordinate. J. Appl. Crystallogr. 28, 768–773 (1995).
Lanza, D.C. et al. Human FEZ1 has characteristics of a natively unfolded protein and dimerizes in solution. Proteins 74, 104–121 (2009).
Petoukhov, M.V. & Svergun, D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 (2005).
Volkov, V.V. & Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003).
Kozin, P.V. & Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 (2001).
Acknowledgements
This work was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; Grants 2006/54878-3, 2007/55930-1, 2007/59442-1, 2008/53519-5, 2008/57805-2, 2010/02628-9) and Conselho Nacional de Pesquisa (CNPq: Grants 304366/2009-9, 475158/2010-5, 573672/2008-3, 559698/2009-7).
Author information
Authors and Affiliations
Contributions
A.M.S., D.S. and K.G.F. conceived the research and designed experiments. A.M.S., D.S., A.C.C., M.B.M.P. and T.M.M. conducted experiments. A.M.S. and C.F.M.Z.C. designed and performed mutational experiments. A.M.S. and A.C.M.F. performed affinity experiments. A.M.S., J.C.S. and I.L.T. performed and analyzed SAXS experiments. A.M.S., M.F. and F.C.G. performed crosslinking and MS experiments and analysis. A.M.S., P.S.L.O. and S.H.P.O. performed molecular modeling and docking. A.M.S., D.S., J.X.N. and K.G.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 2986 kb)
Rights and permissions
About this article
Cite this article
Santos, A., Schechtman, D., Cardoso, A. et al. FERM domain interaction with myosin negatively regulates FAK in cardiomyocyte hypertrophy. Nat Chem Biol 8, 102–110 (2012). https://doi.org/10.1038/nchembio.717
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.717
This article is cited by
-
The roles of nuclear focal adhesion kinase (FAK) on Cancer: a focused review
Journal of Experimental & Clinical Cancer Research (2019)
-
αB-crystallin interacts with and prevents stress-activated proteolysis of focal adhesion kinase by calpain in cardiomyocytes
Nature Communications (2014)
-
Imidate-Based Cross-Linkers for Structural Proteomics: Increased Charge of Protein and Peptide Ions and CID and ECD Fragmentation Studies
Journal of the American Society for Mass Spectrometry (2014)