Abstract
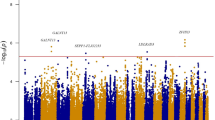
To identify previously unknown genetic loci associated with fasting glucose concentrations, we examined the leading association signals in ten genome-wide association scans involving a total of 36,610 individuals of European descent. Variants in the gene encoding melatonin receptor 1B (MTNR1B) were consistently associated with fasting glucose across all ten studies. The strongest signal was observed at rs10830963, where each G allele (frequency 0.30 in HapMap CEU) was associated with an increase of 0.07 (95% CI = 0.06–0.08) mmol/l in fasting glucose levels (P = 3.2 × 10−50) and reduced beta-cell function as measured by homeostasis model assessment (HOMA-B, P = 1.1 × 10−15). The same allele was associated with an increased risk of type 2 diabetes (odds ratio = 1.09 (1.05–1.12), per G allele P = 3.3 × 10−7) in a meta-analysis of 13 case-control studies totaling 18,236 cases and 64,453 controls. Our analyses also confirm previous associations of fasting glucose with variants at the G6PC2 (rs560887, P = 1.1 × 10−57) and GCK (rs4607517, P = 1.0 × 10−25) loci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Xiang, A.H. et al. Coordinate changes in plasma glucose and pancreatic beta-cell function in Latino women at high risk for type 2 diabetes. Diabetes 55, 1074–1079 (2006).
Mason, C.C., Hanson, R.L. & Knowler, W.C. Progression to type 2 diabetes characterized by moderate then rapid glucose increases. Diabetes 56, 2054–2061 (2007).
Watanabe, R.M. et al. Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Finland-United States Investigation of NIDDM Genetics (FUSION) Study investigators. Hum. Hered. 49, 159–168 (1999).
Weedon, M.N. et al. A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am. J. Hum. Genet. 79, 991–1001 (2006).
Bouatia-Naji, N. et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320, 1085–1088 (2008).
Chen, W.M. et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J. Clin. Invest. 118, 2620–2628 (2008).
Saxena, R. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Vaxillaire, M. et al. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes 57, 244–254 (2008).
Orho-Melander, M. et al. A common missense variant in the glucokinase regulatory protein gene (GCKR) is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 57, 3112–3121 (2008).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008).
Scott, L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Steinthorsdottir, V. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 39, 770–775 (2007).
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Matthews, D.R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Hofman, A. et al. The Rotterdam Study: objectives and design update. Eur. J. Epidemiol. 22, 819–829 (2007).
Herder, C. et al. Variants of the PPARG, IGF2BP2, CDKAL1, HHEX, and TCF7L2 genes confer risk of type 2 diabetes independently of BMI in the German KORA Studies. Horm. Metab. Res. 40, 722–726 (2008).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Myers, A.J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–1499 (2007).
Schadt, E.E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008).
Reppert, S.M. et al. Molecular characterization of a second melatonin receptor expressed in human retina and brain: the Mel1b melatonin receptor. Proc. Natl. Acad. Sci. USA 92, 8734–8738 (1995).
Su, A.I. et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99, 4465–4470 (2002).
Ramracheya, R.D. et al. Function and expression of melatonin receptors on human pancreatic islets. J. Pineal Res. 44, 273–279 (2008).
Stumpf, I., Muhlbauer, E. & Peschke, E. Involvement of the cGMP pathway in mediating the insulin-inhibitory effect of melatonin in pancreatic beta-cells. J. Pineal Res. 45, 318–327 (2008).
Boden, G., Ruiz, J., Urbain, J.L. & Chen, X. Evidence for a circadian rhythm of insulin secretion. Am. J. Physiol. 271, E246–E252 (1996).
Spiegel, K., Leproult, R. & Van, C.E. Impact of sleep debt on metabolic and endocrine function. Lancet 354, 1435–1439 (1999).
Turek, F.W. et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005).
Peschke, E. et al. Melatonin and type 2 diabetes - a possible link? J. Pineal Res. 42, 350–358 (2007).
Acknowledgements
The authors would like to thank the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, as well as genotyping and analysis of the GWA data. They would also like to acknowledge those who agreed to participate in these studies. Major funding for the work described in this paper comes from Academy of Finland (124243); the Administration of Lanusei, Ilbono, Arzana and Elini (Sardinia, Italy); American Diabetes Association (1-05-RA-140); the Center for Inherited Disease Research; Clinical Research Institute (HUCH); Diabetes UK; the European Bioinformatics Institute; the European Commission (contracts LSHM-CT-2006-037197, LSHM-CT-2003-503041, QLK6-CT-2002-02629, QLG2-CT-2002-01254, HEALTH-F4-2007-201413, LSHG-CT-2004-512066, QLRT-2001-01254, LSHG-CT-2004-518153); the Faculty of Biology and Medicine of Lausanne; Finnish Diabetes Research Foundation; Folkhalsan Research Foundation; Foundation of the NIH (GAIN initiative); German Federal Ministry of Education and Research; German Federal Ministry of Health and Social Security; German National Genome Research Network; GlaxoSmithKline; GSF-National Research Center for Environment and Health; LMUinnovativ; Ministry of Science and Research of the State North-Rhine Westphalia; Municipality of Rotterdam; US National Institutes of Health (HG-02651, HL-084729, HL-087679, HC-25195, N02-HL-6-4278, DK-078616, DK-080140, DK-065978, RR-163736, MH059160, DK069922, DA-021519, DK-062370, DK-072193, US National Human Genome Research Institute intramural project HG-000024; and the Intramural Program of the National Institute on Aging); the UK National Institute for Health Research (Oxford Biomedical Research Centre and Guys and St. Thomas' Biomedical Research Centre); the Netherlands Ministry of Education, Culture and Science; the Netherlands Ministry of Health, Welfare and Sports; Novartis; NWO (904-61-090, 904-61-193, 480-04-004, 400-05-717); NWOGenomics; NWOInvestments; Research Institute for Diseases in the Elderly (RIDE); Sigrid Juselius Foundation; Spinozapremie; Swedish Research Council (349-2006-237); UK Medical Research Council (G0500539, G0000649, G016121); UK National Health Services Research and Development; the Wellcome Trust (including intramural support for the Wellcome Trust Sanger Institute, GR069224, Strategic Awards 076113 and 083948, Biomedical Collections Grant GR072960); and ZonMw (10-000-1002). A full list of acknowledgments is provided in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
Project management: DFS: R.N.B., A. Cao, F.S.C., K.L.M., J.T., D.S., M.U., E.L., L.C.G., M. Boehnke, G.R.A.; ENGAGE: P.E., A.H., J.H.S., H.-E.W., G. Willemsen, D.I.B., B.W.J.H.P., M.R.J., L.P., U.T., C.v.D., K. Stefansson, M.I.M.; FHS: J.D., J.B.M.; GEM: T.D.S., I.B., N.J.W.
Study design: DFS: R.S., V.L., R.N.B., T.A.B., A. Cao, F.S.C., K.L.M., L.J.S., J.T., D.S., M.U., E.L., M. Boehnke, R.M.W., G.R.A.; ENGAGE: L.P., A.H., U.T., C.v.D., K. Stefansson, M.I.M.; FHS: J.D., J.C.F., J.B.M.; GEM: C.L., N.S., R.J.F.L., J.S.B., M. Bochud, D.W., M.S., T.D.S., P.V., G. Waeber, V.M., I.B., N.J.W.
Genome-wide association sampling and genotyping: DFS: M.R.E., L.L.B., A. Cao, L. Crisponi, T.E.H., B.I., U.L., S.N., M.O., A.S., H.M.S., T.T., J.T., M.U., D.A., L.C.G.; ENGAGE: P.E., N.F., A.P., E.S., V.S., A.G.U., J.C.M.W., D.I.B., M.R.J., L.P., U.T., C.v.D., K. Stefansson, M.I.M.; FHS: C.S.F., L.A.C., J.D., J.B.M.; GEM: K.A., A. Cervino, P.D., M.I., O.T.M.
Statistical analysis and informatics: DFS: R.S., A.U.J., S. Sanna, W.-M.C., V.G., P.S., B.F.V., R.M.W.; ENGAGE: I.P., G.T., Y.A., J.-J.H., M. Kaakinen, L. Coin, E.J.C.d.G., A.D., C.H., A.K., M. Krestyaninova, C.M.L., J.R., V.S., E.Z., C.v.D.; FHS: A.K.M., J.D.; GEM: C.L., N.S., S.C.P., E.W., S.H., T.J., N.L., S. Sharp, K. Song, X.Y., J.H.Z.
Replication sampling and genotyping: A.S.F.D., A.H., T.I., M.L., A.D.M., C.N.A.P., W.R., J.S., H.E.W.
MAGIC management committee: I.P., C.L., J.C.F., R.S., N.S., G.T., R.J.F.L., A.U.J., Y.A., E.W., V.L., C.M.L., D.W., D.S., M.S., P.V., G. Waeber., L.C.G., V.M., U.T., M. Boehnke, I.B., J.D., R.M.W., M.I.M., N.J.W., J.B.M., G.R.A.
Writing team: I.P., C.L., J.C.F., R.S., N.S., G.T., M. Boehnke, I.B., C.v.D., J.D., R.M.W., K. Stefansson, M.I.M., N.J.W., J.B.M., G.R.A.
Corresponding authors
Ethics declarations
Competing interests
Dawn Waterworth and Vincent Mooser are full-time employees of GlaxoSmithKline. Inês Barroso owns stock in GlaxoSmithKline and Incyte. Drs. Thorleifsson, Kong, Steinsthorsdottir, Thorsteinsdottir, and Stefansson are employees at deCODE genetics, and own stock or stock options in the company. Dr. Meigs currently has research grants from GlaxoSmithKline and Sanofi-Aventis, and serves on consultancy boards for GlaxoSmithKline, Sanofi-Aventis, Interleukin Genetics, Kalypsis, and Outcomes Sciences. Dr. Florez has received consulting honoraria from Merck and from Publicis Healthcare Communications, an advertising agency for Amylin Pharmaceuticals.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Methods, Supplementary Figure 1 and Supplementary Tables 1–6 (PDF 523 kb)
Rights and permissions
About this article
Cite this article
Prokopenko, I., Langenberg, C., Florez, J. et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet 41, 77–81 (2009). https://doi.org/10.1038/ng.290
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.290
This article is cited by
-
Novel targets for potential therapeutic use in Diabetes mellitus
Diabetology & Metabolic Syndrome (2023)
-
Genetics of circadian rhythms and sleep in human health and disease
Nature Reviews Genetics (2023)
-
A novel combination of sitagliptin and melatonin ameliorates T2D manifestations: studies on experimental diabetic models
Journal of Endocrinological Investigation (2023)
-
Interactions between nocturnal melatonin secretion, metabolism, and sleeping behavior in adolescents with obesity
International Journal of Obesity (2022)
-
Gut microbiota mediate melatonin signalling in association with type 2 diabetes
Diabetologia (2022)